Figures
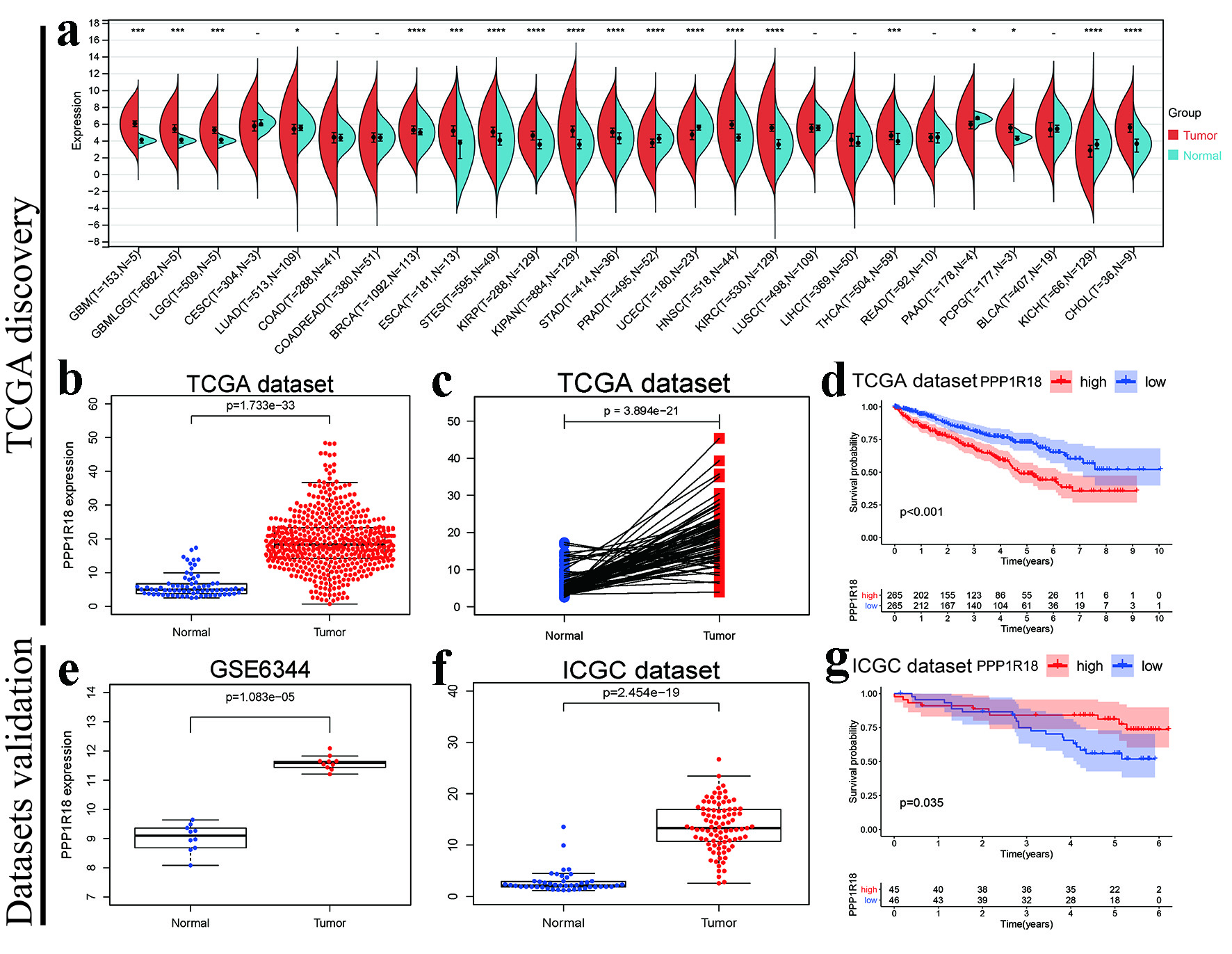
Figure 1. Prognostic roles of PPP1R18 involved in KIRC: (a) PPP1R18 mRNA expression levels in TCGA pan-cancer; (b) Boxplot of PPP1R18 mRNA expression levels in TCGA-KIRC dataset; (c) pairwise boxplot of PPP1R18 mRNA expression levels in TCGA-KIRC dataset; (d) K-M survival analysis of PPP1R18 in TCGA-KIRC dataset; (e) PPP1R18 mRNA expression levels in GSE6344; (f) PPP1R18 mRNA expression levels in ICGC dataset; (g) K-M survival analysis of PPP1R18 in ICGC dataset. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. PPP1R18: protein phosphatase 1 regulatory subunit 18; KIRC: kidney renal clear cell carcinoma; TCGA: The Cancer Genome Atlas; ICGC: International Cancer Genome Consortium.
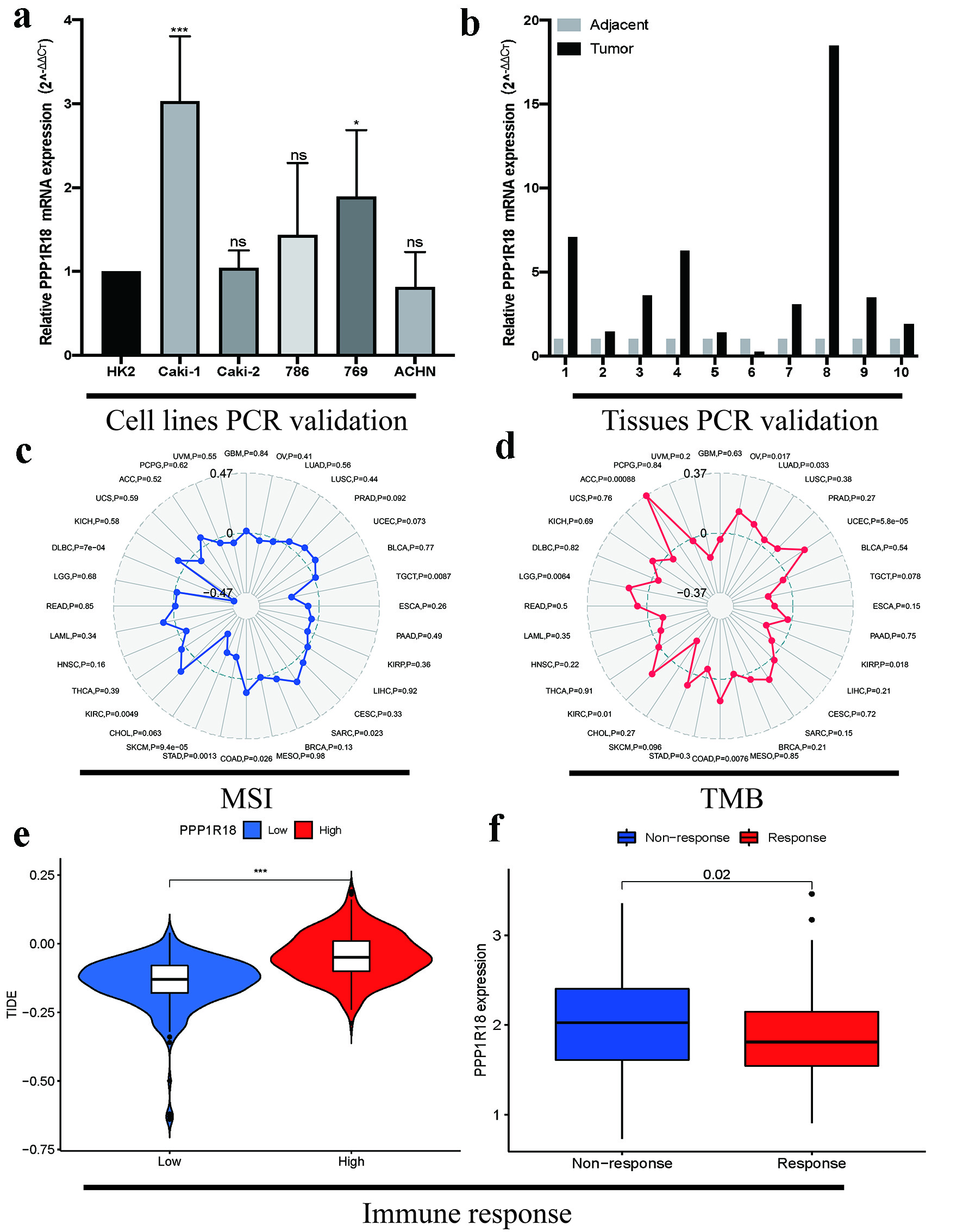
Figure 2. Verification of expression, MSI, TMB and immunotherapy response of PPP1R18 in KIRC: (a) PCR validations in renal cancer cell lines; (b) PCR validations in KIRC tissues; (c) associations between PPP1R18 expression and MSI; (d) associations between PPP1R18 expression and TMB; (e) prediction of immune response by TIDE database; (f) prediction of immune response by IMvigor210 cohort. *P < 0.05; ***P < 0.001. ns: no significance; TMB: tumor mutational burden; MSI: microsatellite instability; PPP1R18: protein phosphatase 1 regulatory subunit 18; KIRC: kidney renal clear cell carcinoma; TIDE: tumor immune dysfunction and exclusion; PCR: polymerase chain reaction.
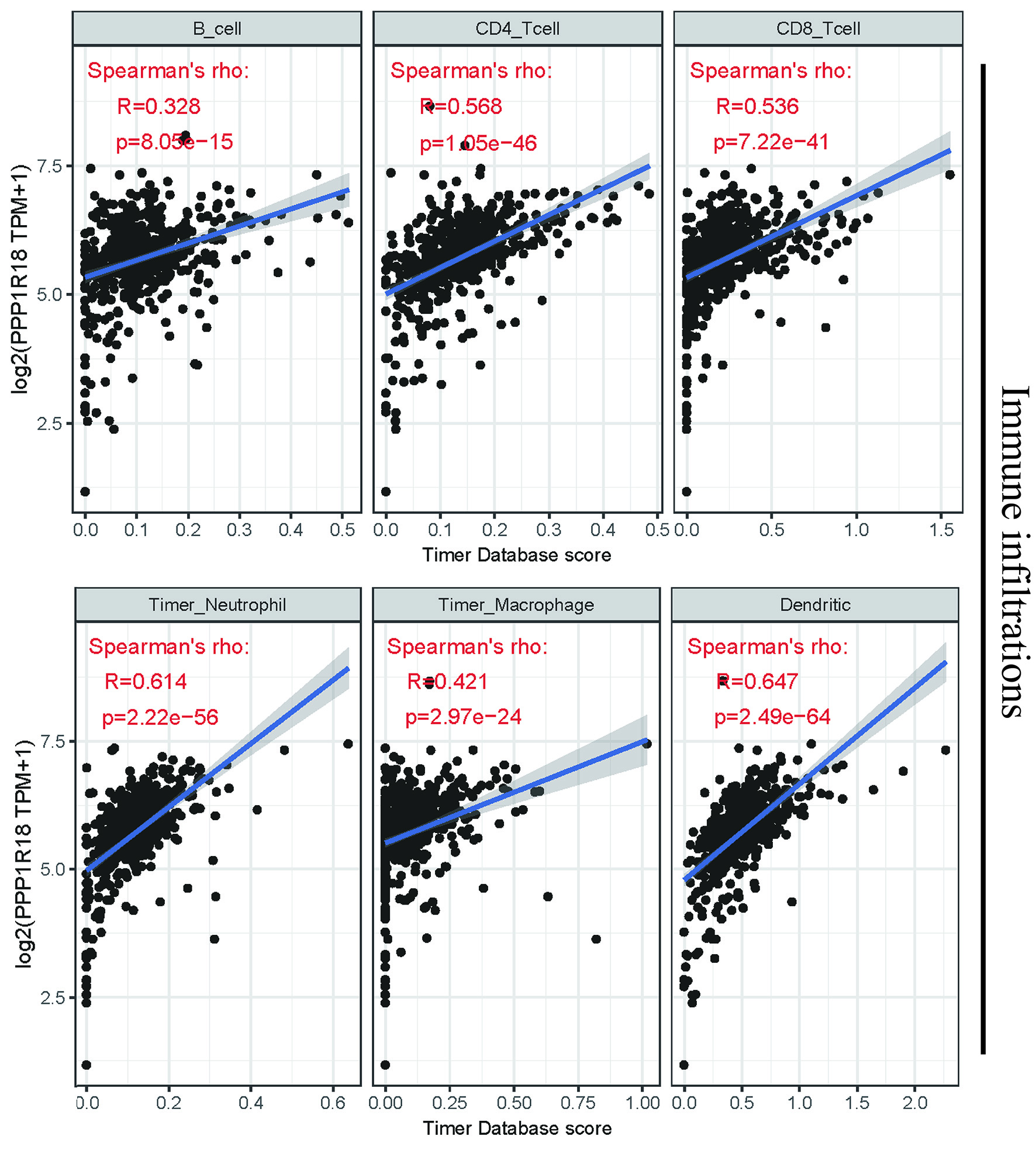
Figure 3. Relationships between PPP1R18 and immune infiltrations in KIRC. PPP1R18: protein phosphatase 1 regulatory subunit 18; KIRC: kidney renal clear cell carcinoma.
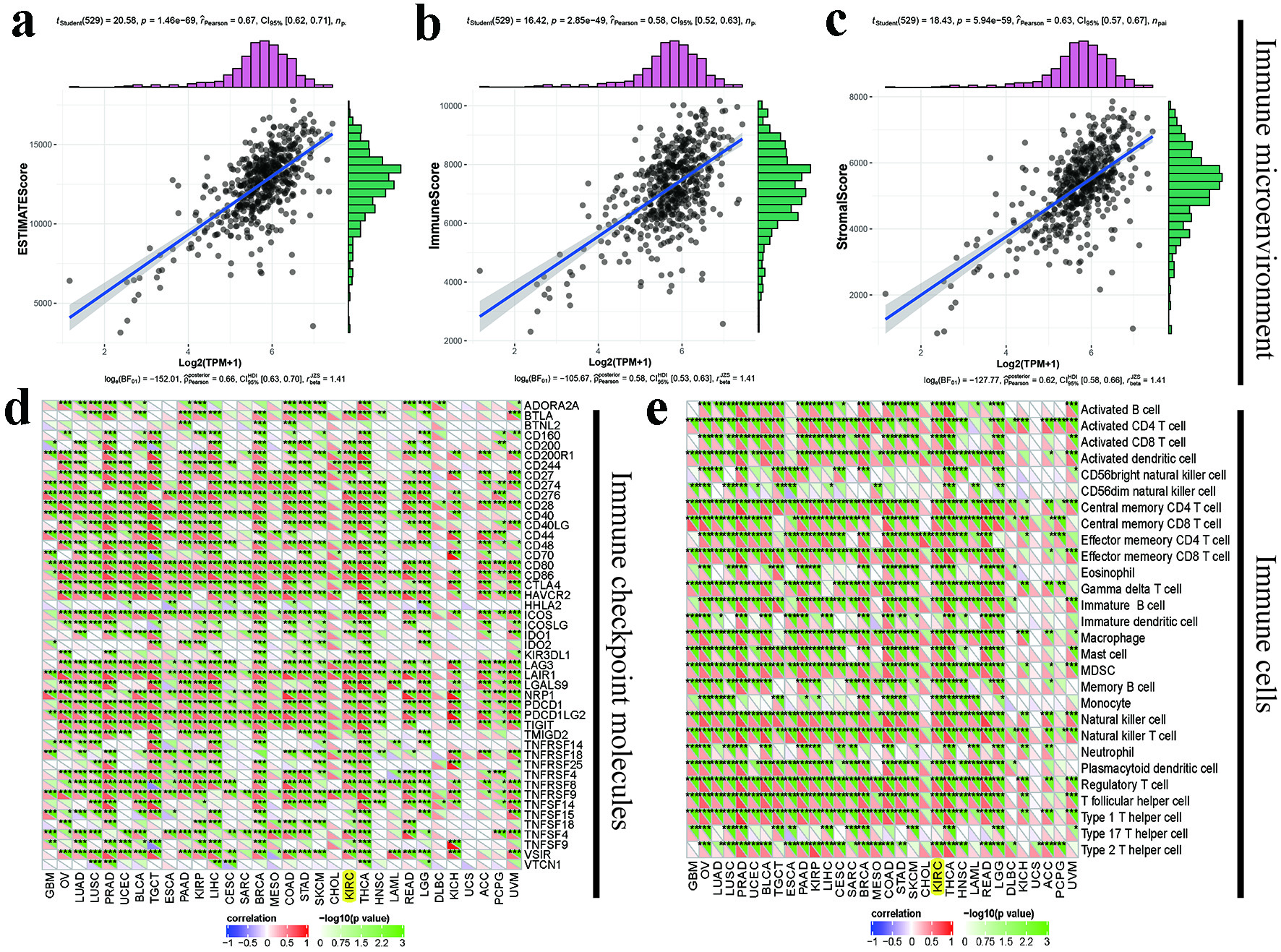
Figure 4. Immunological roles of PPP1R18 involved in KIRC: (a-c) relationships between PPP1R18 and tumor immune microenvironment in KIRC; (d) relationships between PPP1R18 and immune checkpoint molecules; (e) relationships between PPP1R18 and immune cells. *P < 0.05; **P < 0.01; ***P < 0.001. PPP1R18: protein phosphatase 1 regulatory subunit 18; KIRC: kidney renal clear cell carcinoma.
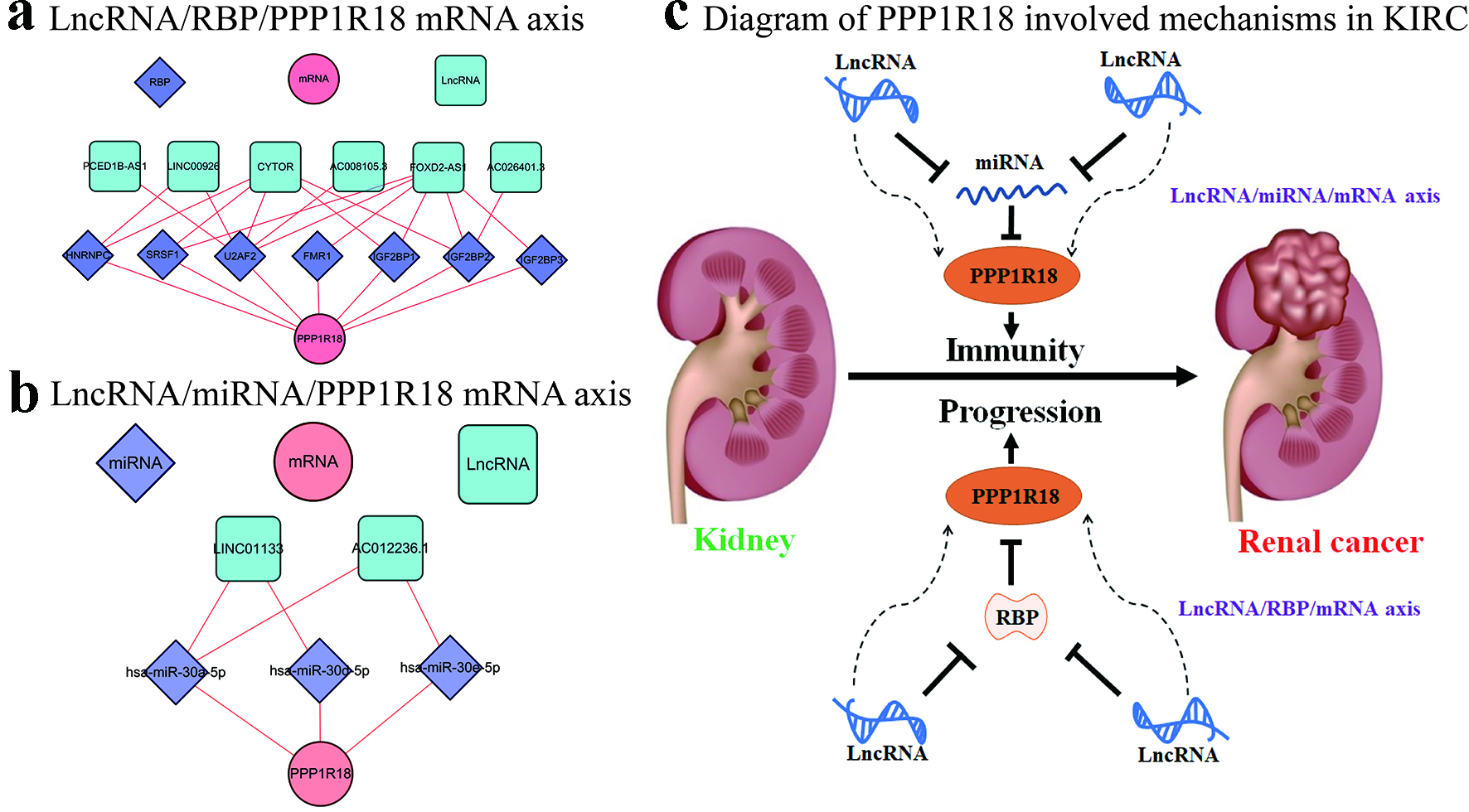
Figure 5. Potential mechanisms of PPP1R18 involved in KIRC: (a) LncRNA/RBP/PPP1R18 mRNA networks in KIRC; (b) LncRNA/miRNA/PPP1R18 mRNA networks in KIRC; (c) diagram of PPP1R18 involved mechanisms in KIRC. PPP1R18: protein phosphatase 1 regulatory subunit 18; KIRC: kidney renal clear cell carcinoma; RBP: RNA-binding protein.
Tables
Table 1. Univariate and Multivariate Analyses of PPP1R18 and Several Clinical Factors for OS in TCGA-KIRC Dataset
| Clinical factors | Univariate analysis | Multivariate analysis |
|---|
| HR | HR.95L | HR.95H | P-value | HR | HR.95L | HR.95H | P-value |
|---|
| PPP1R18: protein phosphatase 1 regulatory subunit 18; KIRC: kidney renal clear cell carcinoma; TCGA: The Cancer Genome Atlas; OS: overall survival. |
| Age | 1.033274 | 1.019678 | 1.047052 | 1.28 × 10-6 | 1.039339 | 1.023979 | 1.05493 | 3.79 × 10-7 |
| Gender | 0.933298 | 0.679692 | 1.28153 | 0.669603 | 0.942881 | 0.681707 | 1.304114 | 0.722276 |
| Race | 1.193075 | 0.71596 | 1.988138 | 0.498059 | 1.098543 | 0.640712 | 1.883524 | 0.73261 |
| Grade | 1.966884 | 1.638836 | 2.360598 | 3.70 × 10-13 | 1.367854 | 1.086933 | 1.721381 | 0.00757 |
| Stage | 1.855626 | 1.643637 | 2.094956 | 1.71 × 10-23 | 1.653636 | 1.166588 | 2.344024 | 0.00472 |
| T | 1.997582 | 1.689052 | 2.362469 | 6.29 × 10-16 | 1.075681 | 0.819392 | 1.412131 | 0.599303 |
| M | 2.099647 | 1.660681 | 2.654644 | 5.70 × 10-10 | 0.9157 | 0.484497 | 1.730671 | 0.786276 |
| N | 0.862971 | 0.73887 | 1.007916 | 0.062826 | 0.847414 | 0.721356 | 0.995501 | 0.043922 |
| PPP1R18 | 1.04568 | 1.028042 | 1.06362 | 2.66 × 10-7 | 1.027619 | 1.008643 | 1.046953 | 0.004171 |
Table 2. GSEA Results of PPP1R18 Involved Pathways in TCGA-KIRC Dataset
| MSigDB collection | Gene set name | NES | NOM P-value | FDR Q-value |
|---|
| GSEA: gene set enrichment analysis; PPP1R18: protein phosphatase 1 regulatory subunit 18; KIRC: kidney renal clear cell carcinoma; TCGA: The Cancer Genome Atlas. |
| c2.cp.kegg.v7.1.symbols.gmt | B_CELL_RECEPTOR_SIGNALING_PATHWAY | 2.225 | < 0.001 | 0.001 |
| CHEMOKINE_SIGNALING_PATHWAY | 2.517 | < 0.001 | < 0.001 |
| JAK_STAT_SIGNALING_PATHWAY | 2.407 | < 0.001 | < 0.001 |
| MAPK_SIGNALING_PATHWAY | 2.150 | < 0.001 | 0.003 |
| NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 2.145 | 0.004 | 0.003 |
| NOTCH_SIGNALING_PATHWAY | 2.191 | < 0.001 | 0.002 |
| T_CELL_RECEPTOR_SIGNALING_PATHWAY | 2.297 | < 0.001 | < 0.001 |
| TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 2.210 | 0.006 | 0.002 |
| VEGF_SIGNALING_PATHWAY | 2.032 | < 0.001 | 0.006 |




