Figures
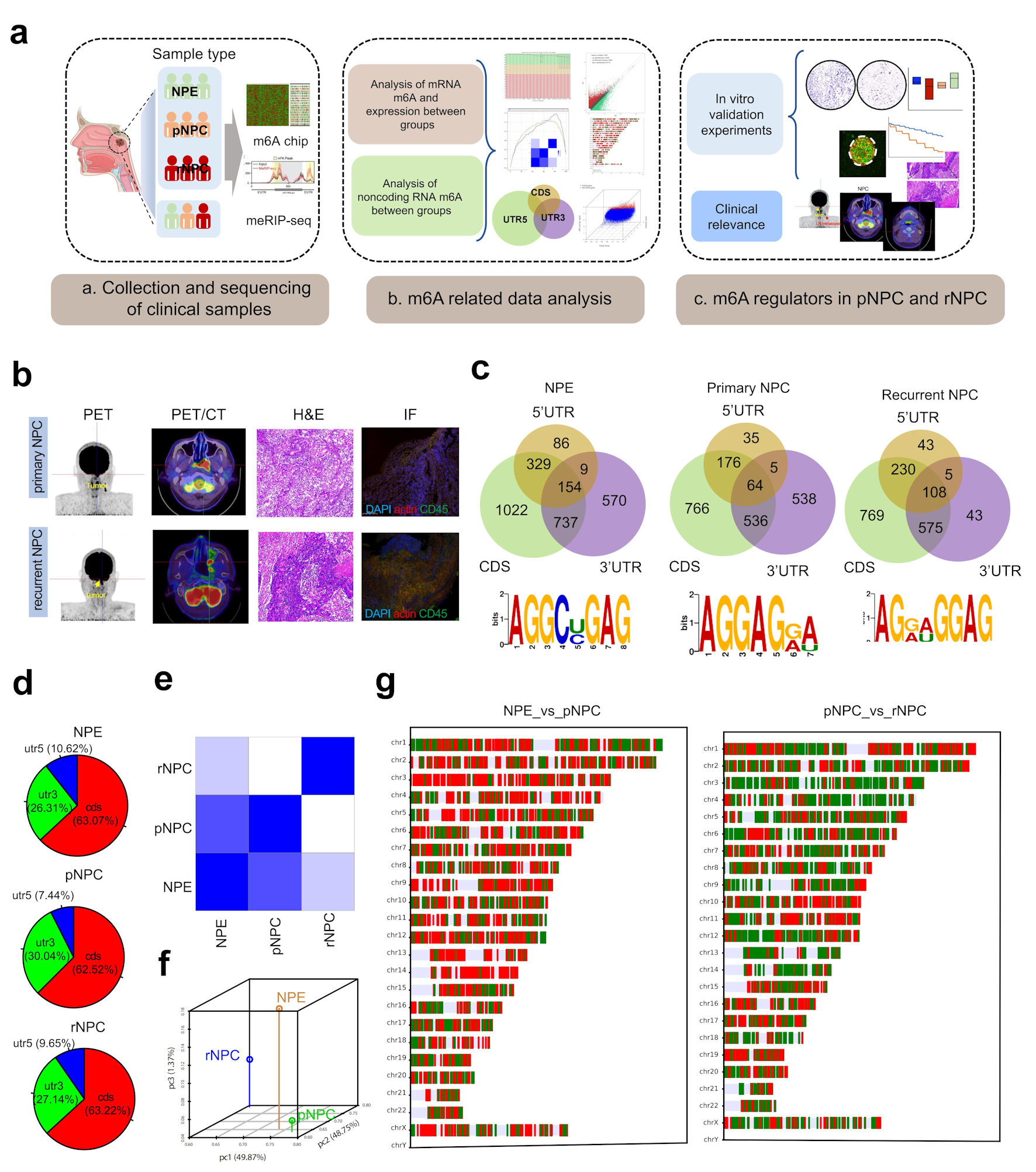
Figure 1. Landscape of RNA m6A modification in NPE, primary and recurrent nasopharyngeal carcinoma. (a) The experimental scheme for discovering and validating m6A modification in NPC. (b) PET/CT and immunohistochemistry (IHC) of representative primary and recurrent NPC patient. (c) The distribution of m6A on mRNA region and the m6A-prone motif in NPC tissues. (d) The distribution of differently methylated mRNA between groups. (e) Heatmap of PCA correlation between NPE, pNPC and rNPC mRNA expression. (f) Dot plot of PCA correlation between NPE, pNPC, and rNPC mRNA expression. (g) Distribution of differentially expressed mRNA molecules on chromosomes; red represents higher expression, blue represents lower repression. CDS: coding sequence; UTR: untranslated region; NPC: nasopharyngeal carcinoma; NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma; PET/CT: positron emission tomography/computed tomography scan; H&E: hematoxylin and eosin; m6A: N6-methyladenosine; IF: immunofluorescence.
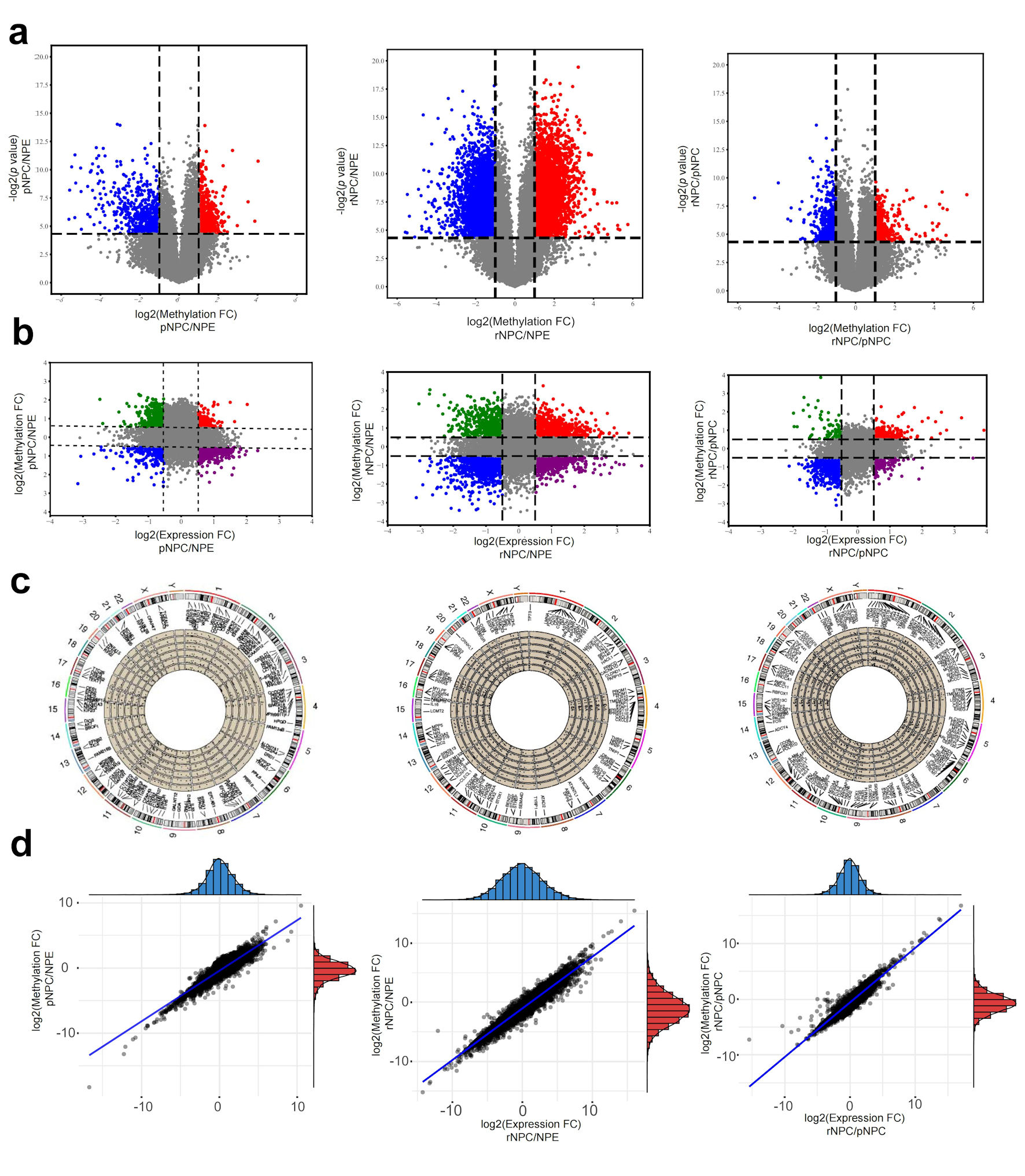
Figure 2. Differentially m6A modification and differentially expressed mRNAs in NPC. (a) Differential m6A modificated mRNA molecules in pNPC/NPE, rNPC/NPE and rNPC/pNPC groups (from left to right). (b) Relationship between mRNA methylation fold change and expression fold change in pNPC/NPE, rNPC/NPE and rNPC/pNPC groups (from left to right). (c) Distribution of differential mRNA molecules on chromosomes in pNPC/NPE, rNPC/NPE, and rNPC/pNPC groups. (d) Relationship between mRNA methylation and expression level in pNPC/NPE, rNPC/NPE, and rNPC/pNPC groups (from left to right). NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma.
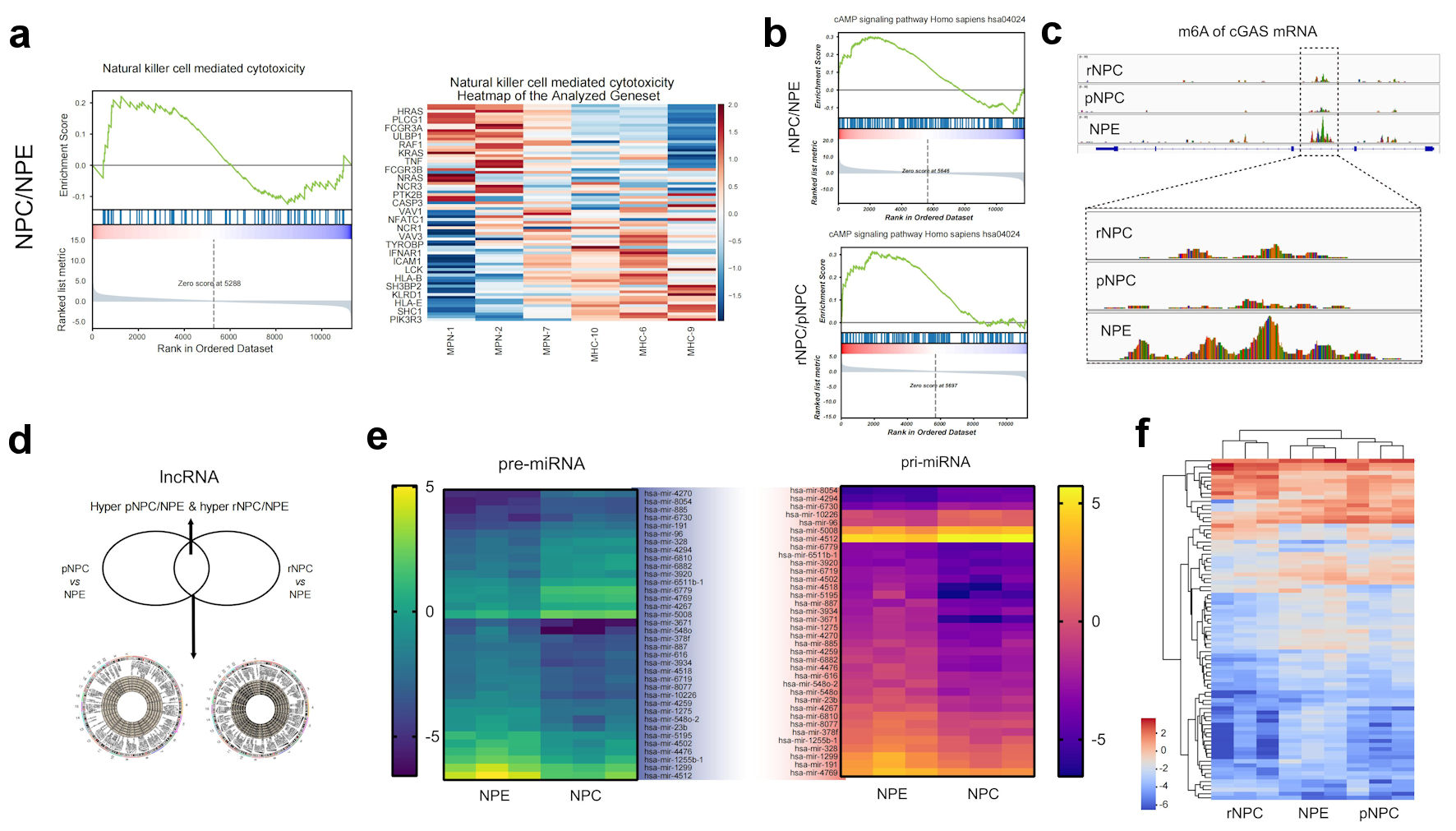
Figure 3. Function analysis of differently methylated mRNA and noncoding RNAs. (a) Represent GSEA enrichment function for differential methylation mRNA molecules in NPC. (b) Represent GSEA enrichment signaling pathways for differential methylation mRNA molecules in NPC. (c) m6A modification details of representative molecule cGAS by meRIP sequencing. (d) Differently methylated lncRNAs in primary NPC and NPE tissues. (e) Heatmap of differently methylated pre-miRNAs and pri-miRNAs in primary NPC and NPE tissues. (f) Heatmap of differently methylated snoRNA in NPC and NPE tissues. NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma; m6A: N6-methyladenosine; lncRNA: long noncoding RNA; me RIP: methylated RNA immunoprecipitation; cGAS: cyclic GMP-AMP synthase; miRNAs: micro RNAs; pre-miRNAs: precursor miRNAs; pri-miRNAs: primary miRNAs.
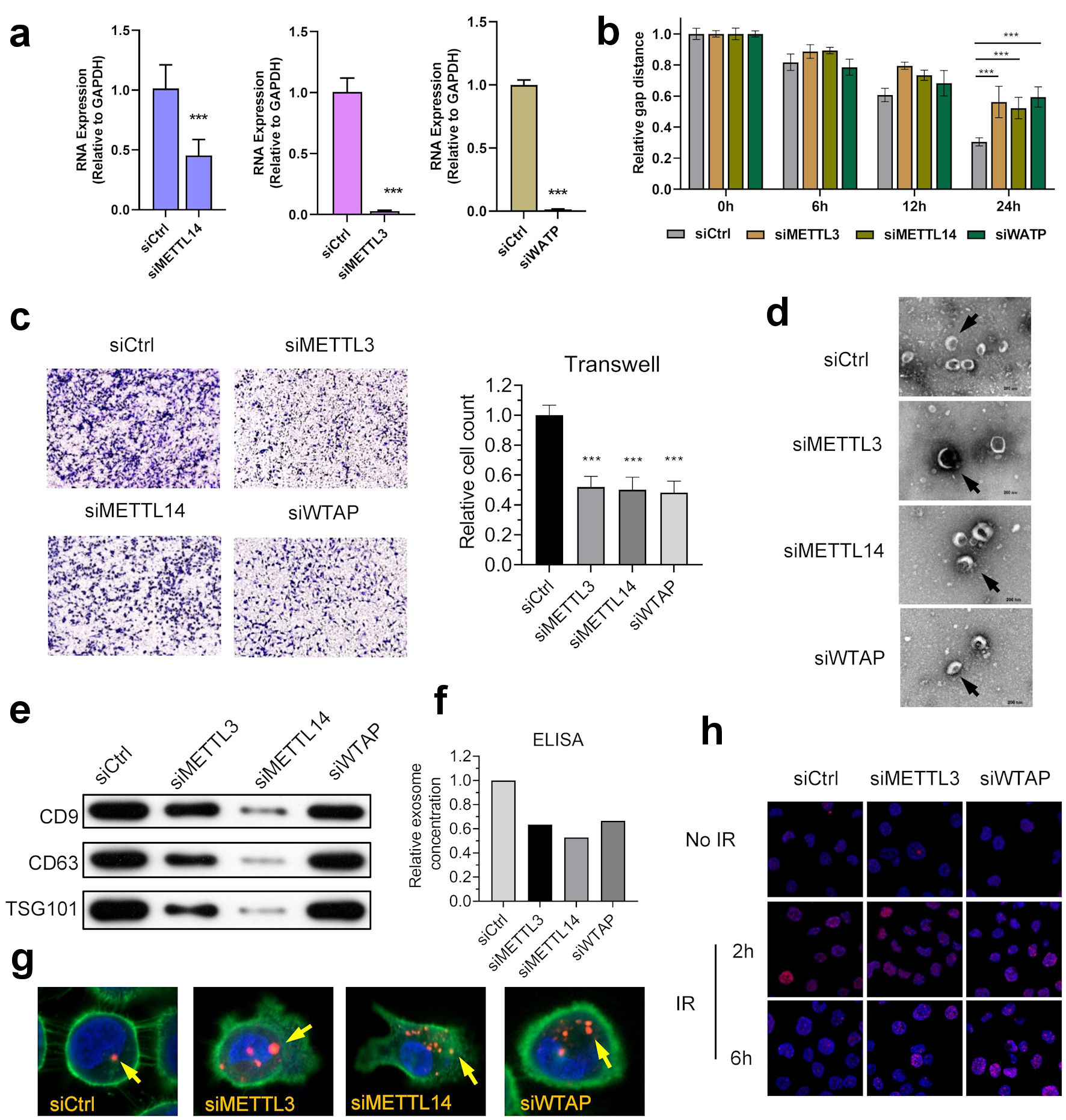
Figure 4. m6A regulators anticipate in the development and progression of NPC. (a) Knocking down of WTAP, METTL3 and METTL14 in NPC cell line HK-1. (b) WTAP, METTL3, and METTL14 promoted the migration ability of HNE2 cells transfected with siRNA assessed using by wound-healing assay. Images were acquired at 0 h to 24 h. Data have been represented as represent with mean ± SD. *P < 0.05; ***P < 0.001. (c) WTAP, METTL3, and METTL14 promoted the invasive ability of S18 cells after knockdown, as assessed using by Transwell® invasion assays. Images were acquired at 24 h and are representative of and presented from three independent experiments. Data have been represented as represent with mean ± SD. **P < 0.01; ***P < 0.001. (d) Representative cryo-electron microscopy images of exosomes in four groups (scale bar = 200 nm). (e) Western blotting analysis of exosome markers CD9, CD63, and TSG101 in four groups. (f) ELISA showed that knockdown of m6A writers would reduce exosome in NPC microenvironment. (g) Immunofluorescence showed that knockdown of m6A writers can induce uptake of exosome in NPC cells. (h) Represent image of γH2AX IRIF in 5-8F cells were quantified after 2 Gy IR at indicated recovery time in siControl, siMETTL3 and siMETTL14 group. METTL3: methyltransferase 3; METTL14: methyltransferase 14; WTAP: WT1 associated protein; ELISA: enzyme-linked immunoassay; IR: ionizing radiation; IRIF: IR-induced foci.
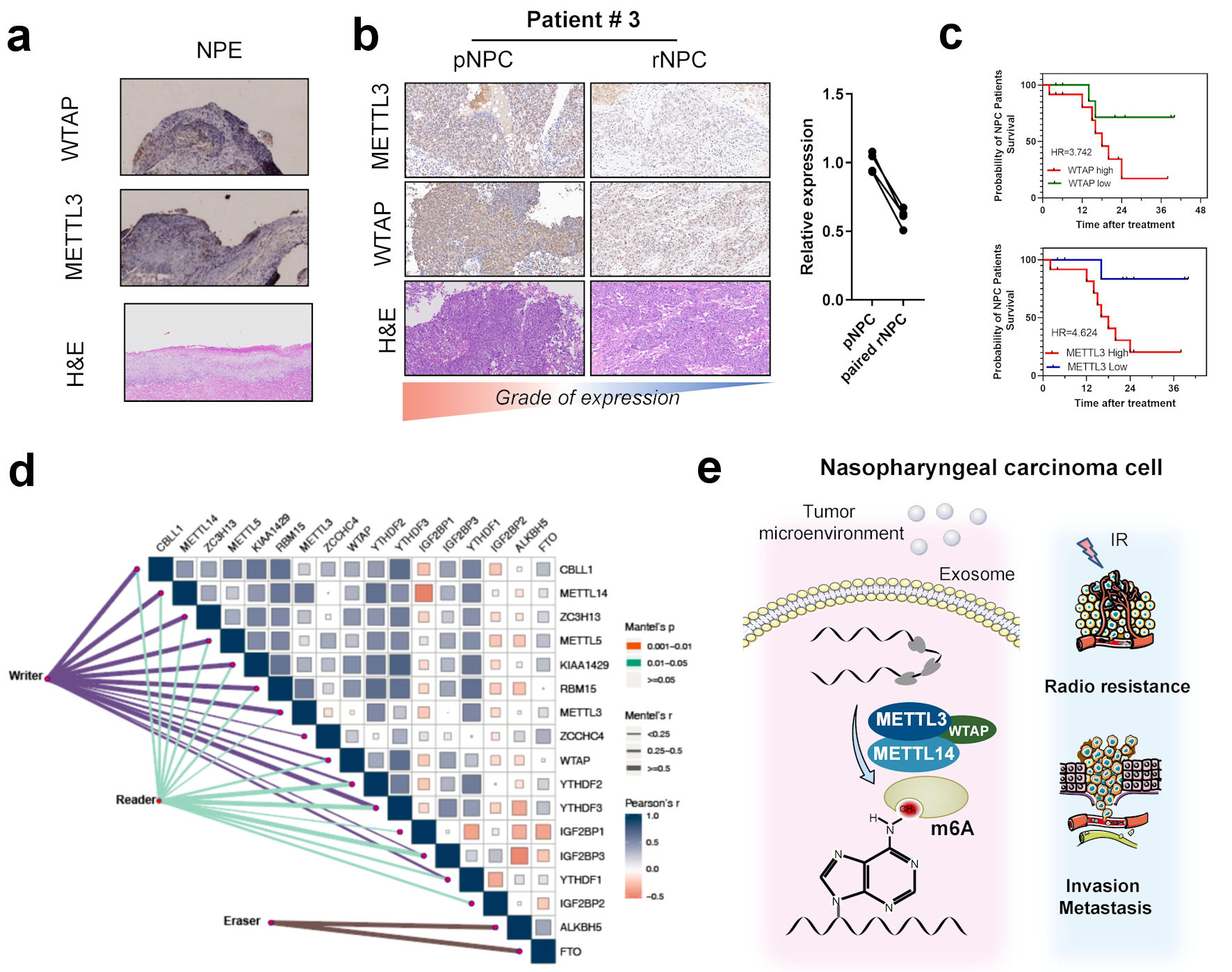
Figure 5. Correlation between m6A regulators with clinical outcomes in nasopharyngeal carcinoma. (a) Immunohistochemistry of m6A writer METTL3, WTAP, and hematoxylin and eosin (H&E) staining in NPE tissues. (b) Immunohistochemistry of m6A writer METTL3, WTAP, and H&E staining in primary NPC and paired recurrent NPC tissues. (c) K-M curve of overall survival in patients with different METTL3 and WTAP expression. (d) Correlation between expression of m6A regulators including eraser reader and writers in head and neck cancer patients’ tissues in TCGA-HNSC cohort. (e) The scheme of the role of m6A and regulators in the occurrence and recurrence of nasopharyngeal carcinoma. METTL3: methyltransferase 3; METTL14: methyltransferase 14; WTAP: WT1 associated protein; NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma; m6A: N6-methyladenosine; TCGA: The Cancer Genome Atlas; HNSC: head and neck squamous cell carcinoma.




