Figures
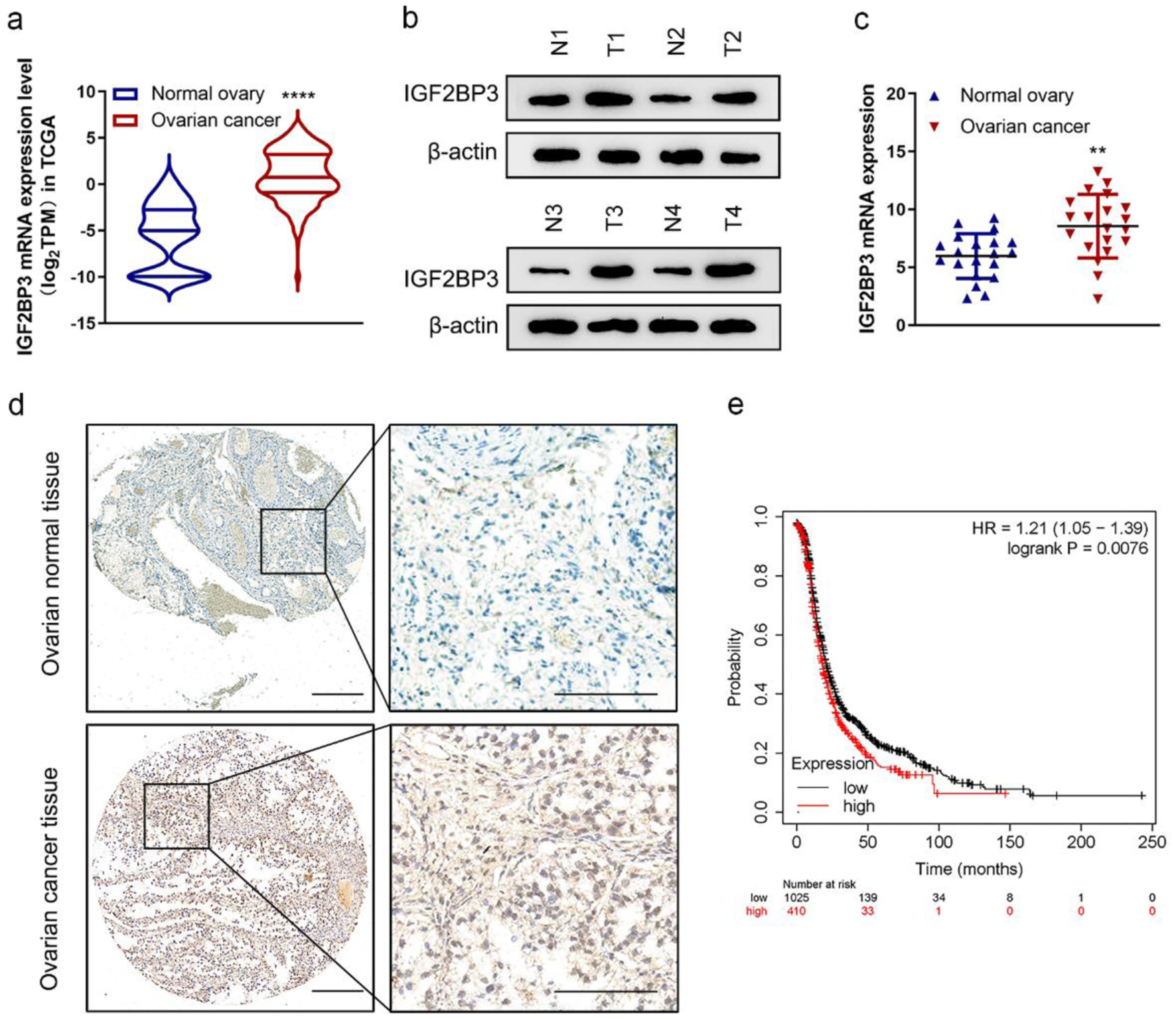
Figure 1. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) expression is increased in ovarian cancer (OC). (a) The mRNA expression of IGF2BP3 in OC (n = 419) tissues versus in the adjacent tissues (n = 88) from The Cancer Genome Atlas (TCGA) data. ****P < 0.0001, OC vs. normal ovary. (b) Representative images of western blot analysis demonstrating the protein level of IGF2BP3 in normal (N) and cancerous (T) ovarian tissues. (c) The expression of IGF2BP3 was analyzed by qRT-PCR and normalized to GAPDH in 20 pairs of normal and cancerous ovarian tissues. (d) Representative immunohistochemistry (IHC) images reflecting the IGF2BP3 expression in OC and normal ovarian tissues. Scale bars: 50 µm (left) and 200 µm (right). (e) Kaplan-Meier curves of progression-free survival (PFS) for OC patients with high (n = 410) and low (n = 1,025) levels of IGF2BP3 (probe ID 203819_s_at).
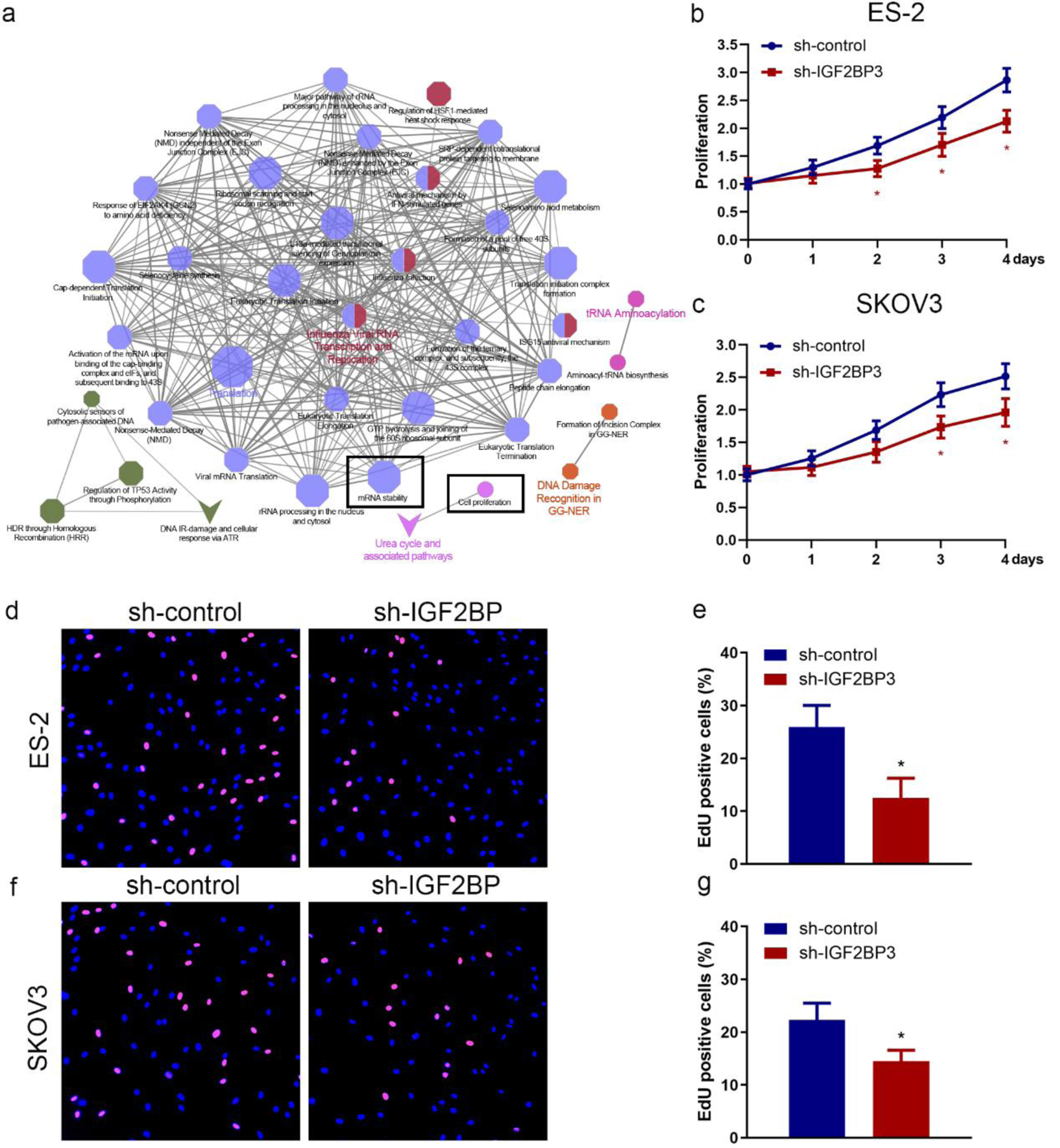
Figure 2. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) knockdown suppresses ovarian cancer (OC) cell proliferation. (a) The pathways impacted by the silencing of IGF2BP3 in OC cells were examined using the clueGO and cluePedia plugins of the cytoscape software. Through this analysis, the cluster of cell proliferation and mRNA stability pathway were found to be enriched and were identified based on statistical analysis. Cell proliferation assay results of MTS assay (b and c) and 5-ethynyl-2′-deoxyuridine (EdU) assay (d-g) evaluating the effect of IGF2BP3 depletion on the growth in ES-2 and SKOV3 cells (*P < 0.05, sh-IGF2BP3 vs. sh-control). (d-g) Representative images (d and f) and quantified results (e and g) of EdU assay determining the effect of IGF2BP3 depletion on the growth in ES-2 (upper panel) and SKOV3 (lower panel) cells. Error bars represent mean ± SD from three experiments. **P < 0.01, sh-IGF2BP3 vs. sh-control.
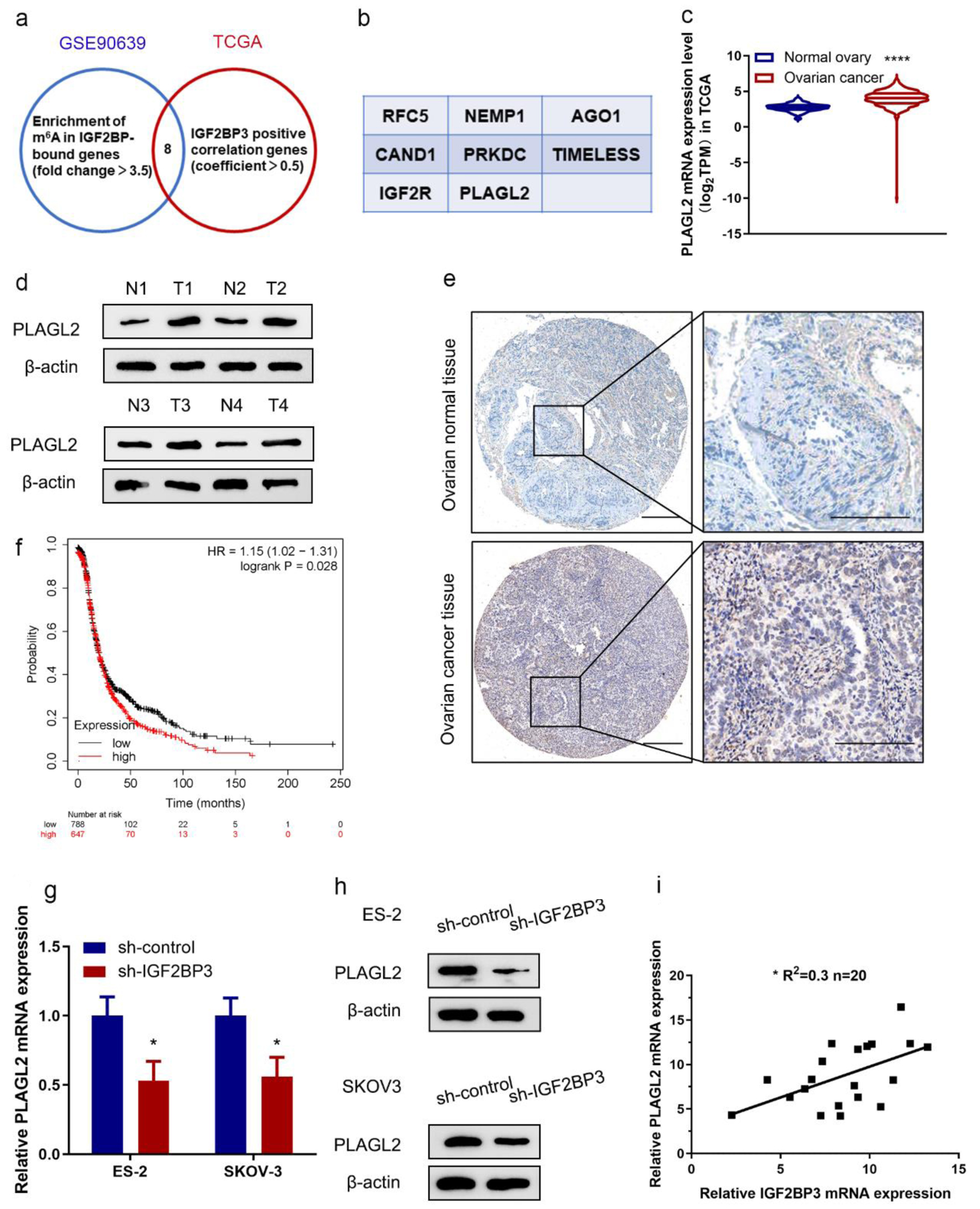
Figure 3. PLAGL2 is a downstream target of insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) in ovarian cancer (OC). (a) Venn diagram showing genes from indicated datasets were positively correlated with IGF2BP3 and/or downregulated in IGF2BP3-deficient OC cells. (b) Genes from the Venn diagram that are both correlated with IGF2BP3 and downregulated in IGF2BP3-dificient OC cells. (c) The mRNA expression of PLAGL2 in OC tissues (n = 419) versus in the adjacent tissues (n = 88) from The Cancer Genome Atlas (TCGA) data. ****P < 0.0001, OC vs. normal ovary. (d) Western blot analysis of PLAGL2 expression in paired normal and cancerous ovarian tissues. (e) Representative immunohistochemistry (IHC) images showing PLAGL2 expression in cancerous and normal ovarian tissues. Scale bars: 50 µm (left) and 200 µm (right). (f) Kaplan-Meier curves of progression-free survival (PFS) for OC patients with high (n = 647) and low (n = 788) levels of PLAGL2 (probe ID 202925_at). (g and h) Results of RT-qPCR (h) and western blot analysis (h) detecting the expression of PLAGL2 in ES-2 and SKOV3 cells transduced with or without IGF2BP3-targeting shRNAs. (i) Paired 20 normal and cancerous ovarian for qRT-PCR assay. Pearson’s correlation analysis of the relative expression levels of IGF2BP3 and PLAGL2.
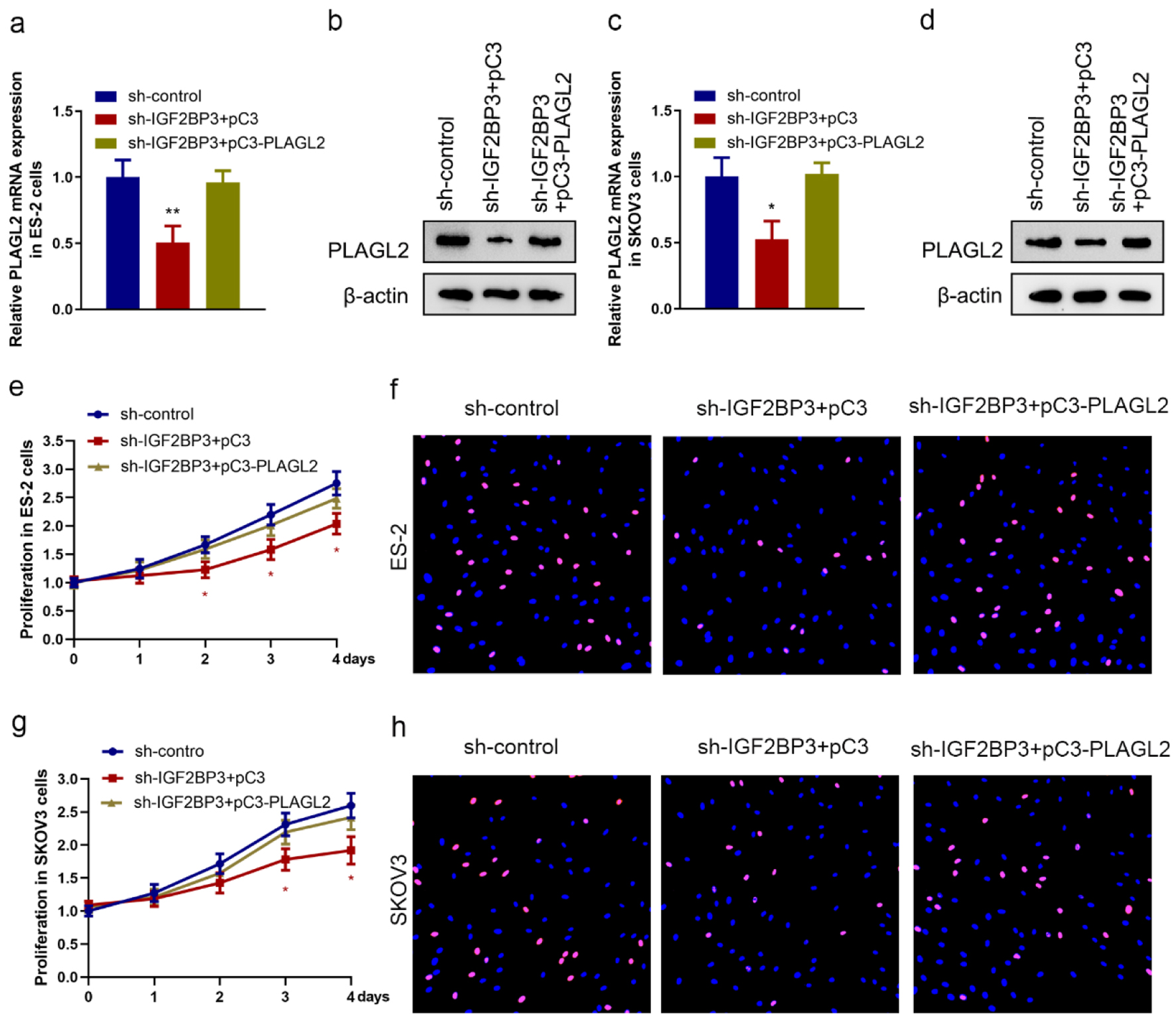
Figure 4. Silencing insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) attenuates ovarian cancer (OC) cell proliferation in a PLAGL2-dependent manner. (a, b, c, and d) The expression of PLAGL2 in IGF2BP3-depleted and PLAGL2 overexpressing cells was assessed using RT-qPCR and western blot analysis in ES-2 cells (a and b) and SKOV3 cells (c and d). (*P < 0.05, **P < 0.01). Cell proliferation results of MTS assays (e and g) and EdU (f and h) assay using ES-2 cells and SKOV3 cells under indicated conditions (IGF2BP3-depleted and PLAGL2 overexpressing cells) (*P < 0.05).
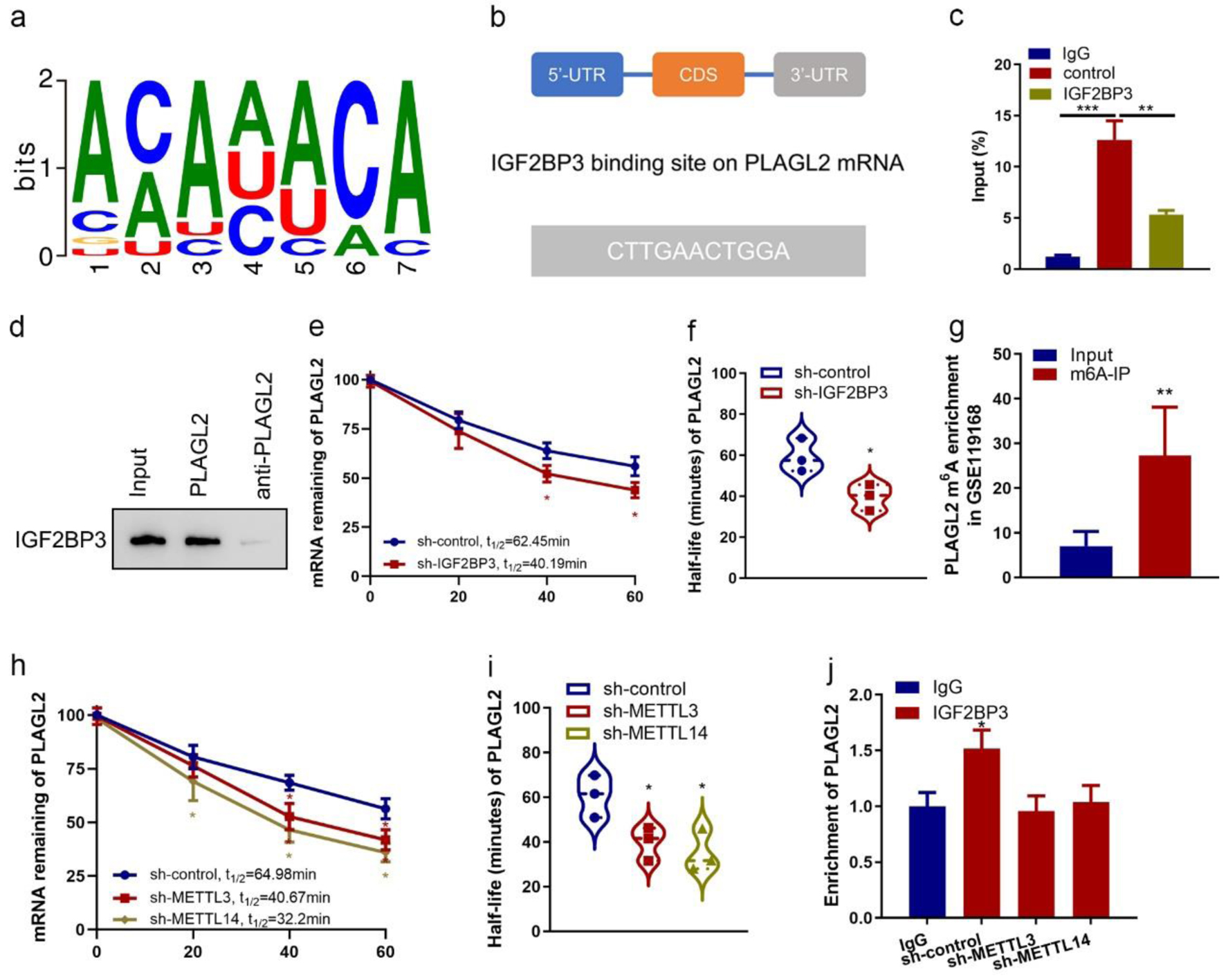
Figure 5. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) binds with PLAGL2 mRNA and regulates its stability. (a) Consensus sequences of IGF2BP3-binding sites by motif prediction. (b) The binding site of IGF2BP3 on PLAGL2 mRNA was predicted. The 3′-UTR sequence with potential binding sites (blue) is shown. (c) Enrichment of PLAGL2 in ES-2 cells performed RIP of IGF2BP3 and control using anti-flag antibody from nuclear extractions and identified the associated PLAGL2 mRNAs by RT-qPCR. (***P < 0.001, **P < 0.01). (d) The cell lysate of ES-2 cells was utilized in an RNA pulldown assay employing biotin-labeled PLAGL2 and PLAGL2 antisense, followed by western blotting analysis. (e) Result of the mRNA half-life measurement showing the decaying of PLAGL2 mRNA in parental and IGF2BP3-depleted cells (*P < 0.05, sh-IGF2BP3 vs. sh-control). (f) Half-life values in the same treatment groups. (g) Results of bioinformatics analysis of online dataset (GSE90639) revealing the enrichment of PLAGL2 m6A-modified RNA by IGF2BP3 (***P < 0.001, m6A probe vs. unmethylated probe). (h) Result of the mRNA half-life measurement showing the decaying of PLAGL2 mRNA in parental and METTL3- or METTL14-depleted cells (*P < 0.05, vs. sh-control). (i) Half-life values in the same treatment groups. (j) Enrichment of PLAGL2 in parental and METTL3- or METTL14-depleted ES-2 cells performed RIP of IGF2BP3 and control using anti-flag antibody and identified the associated PLAGL2 mRNAs by RT-qPCR. Input percentage is shown (*P < 0.05).
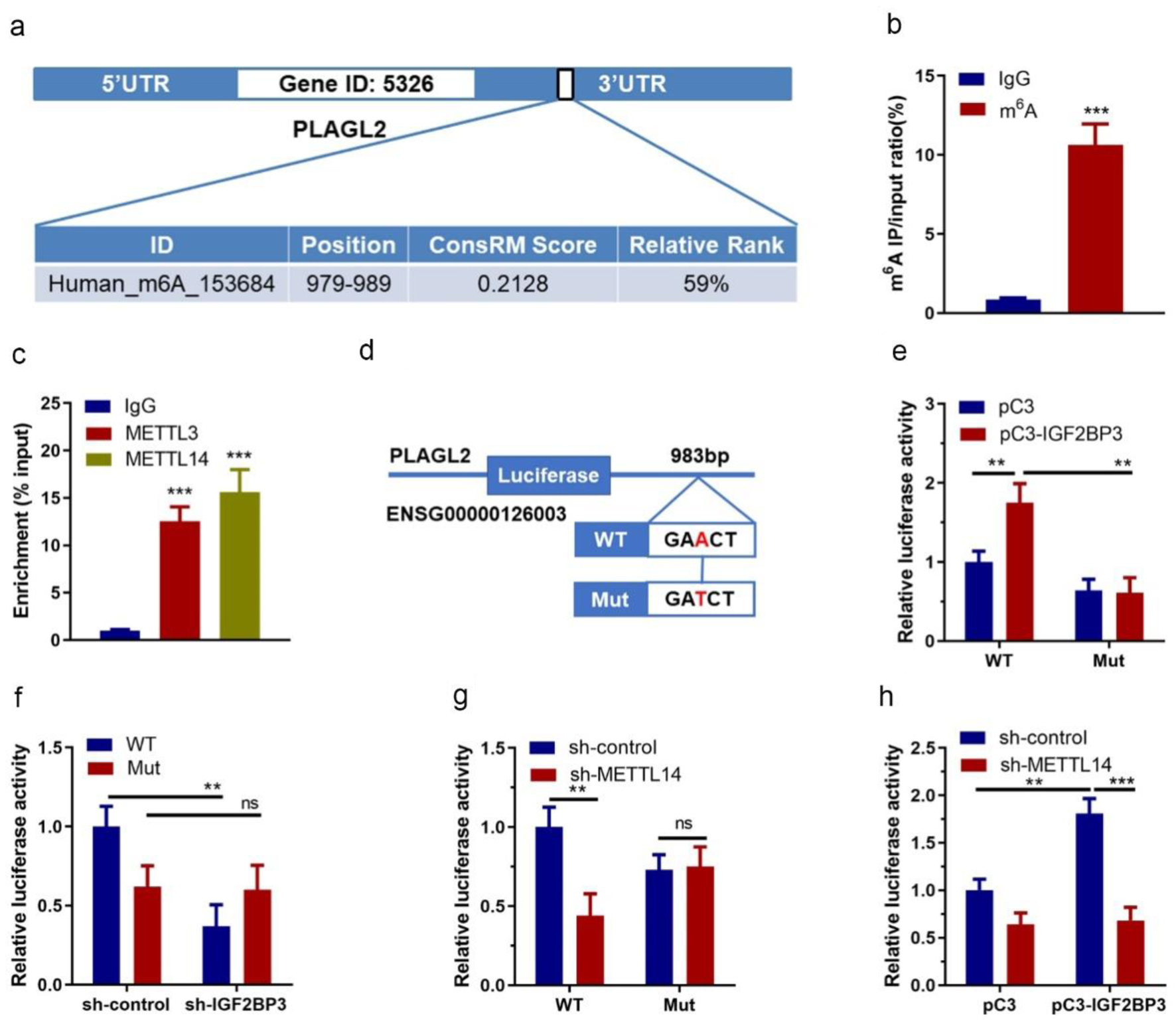
Figure 6. Identification of m6A methylation between PLAGL2 mRNA and insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3). (a) The prediction of the PLAGL2 m6A RNA methylation site using online tool (m6Avar). (b) Results of PLAGL2-specific m6A RT-qPCR assay (*** P<0.001, m6A vs. IgG). (c) Results of RIP assay showing the interactions between PLAGL2 and METTL3/METTL14 (***P < 0.001, vs. IgG). (d) A schematic diagram showing the wildtype and mutated methylation site on PLAGL2 3'-UTR. (e) Results of dual luciferase assay in ES-2 cells co-transfected with vechicle vector or IGF2BP3-overexpressing vector and reporter plasmid containing wildtype or mutant PLAGL2 3'-UTR (*P < 0.05, **P < 0.01, vs. pC3). (f) Results of dual luciferase assay in ES-2 cells with or without IGF2BP3 knockdown transfected with reporter plasmid containing wildtype or mutant PLAGL2 3'-UTR (*P < 0.05, vs. WT sh-control). (g) Results of dual luciferase assay in ES-2 cells co-transfected with vechicle vector or METTL14-targeting shRNA and reporter plasmid containing wildtype or mutant PLAGL2 3'-UTR (*P < 0.05, vs. WT sh-control). (h) Relative luciferase activity of PLAGL2 wildtype 3'-UTR or mutation in METTL14 stable knockdown or control ES-2 cells with ectopic expression of IGF2BP3.
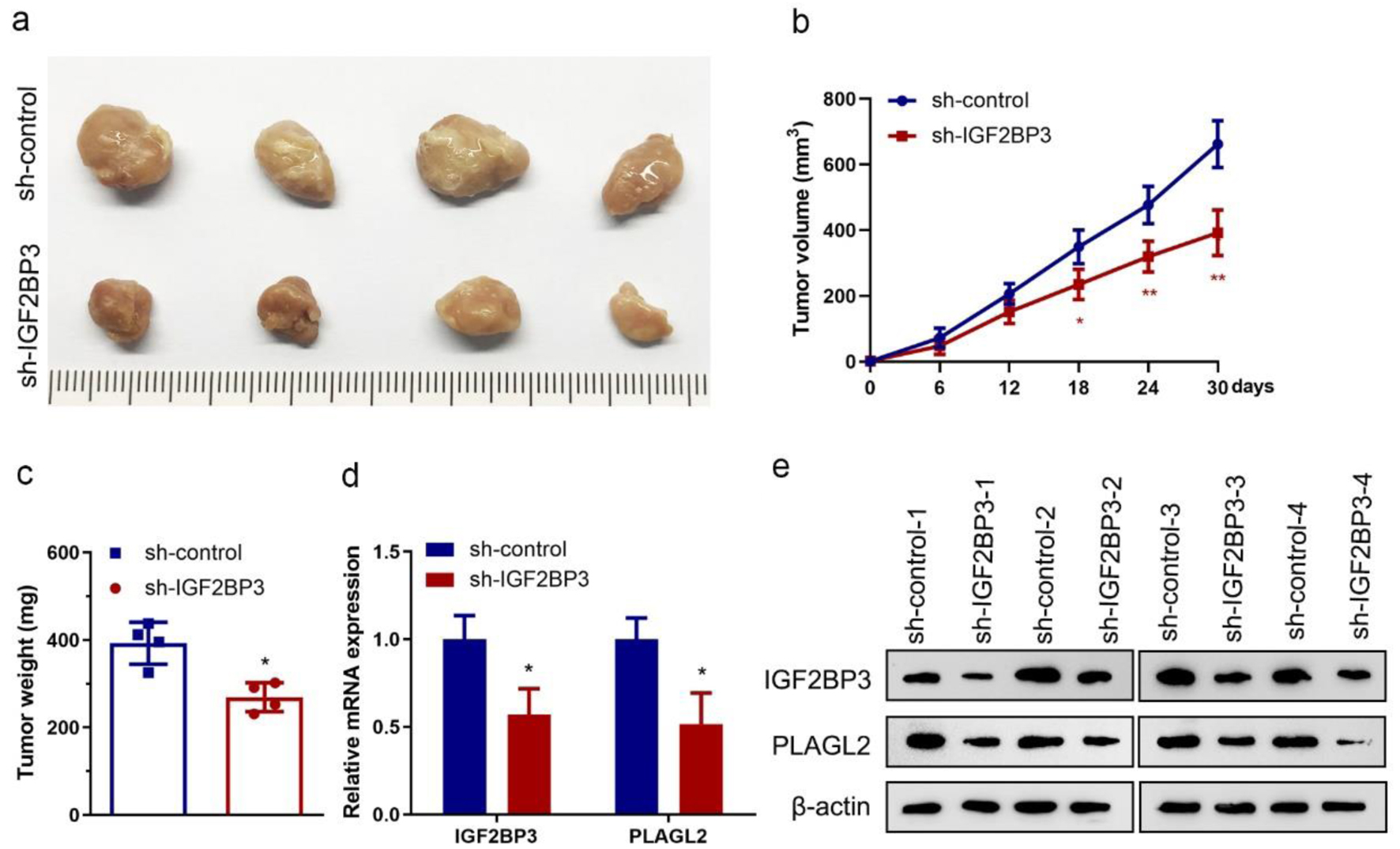
Figure 7. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) knockdown impairs tumor growth in vivo. (a) Images showing tumors harvested from each group. (b) Plotted data depicting tumor growth curves in different groups (n = 4) (*P < 0.05, **P < 0.01, vs. sh-control). (c) Analyzed data showing the individual tumor weight from each group (n = 4) (*P < 0.05, vs. sh-control). (d and e) Results of RT-qPCR (d) and representative images of western blot analysis (e) showing the expression of IGF2BP3 and PLAGL2 in harvested xenograft tumors (*P < 0.05, vs. sh-control).
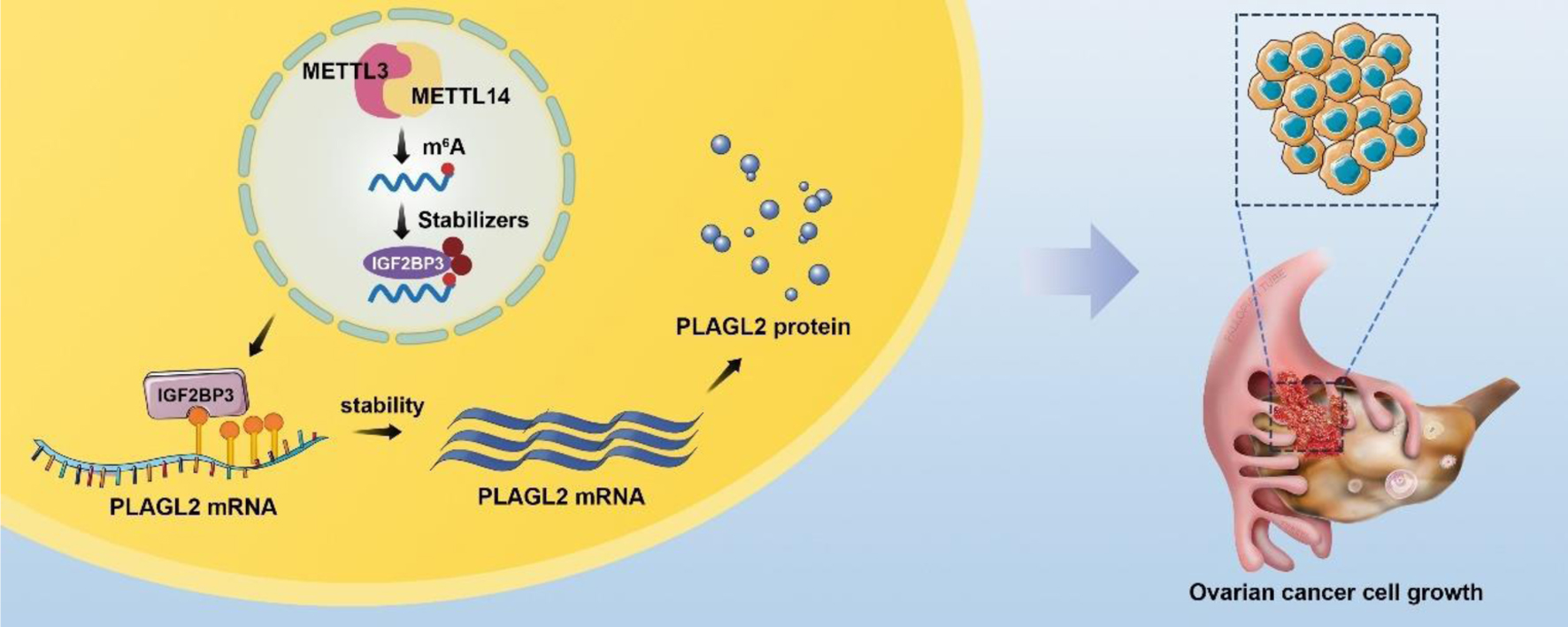
Figure 8. A schematic diagram for the mechanisms of insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) knockdown on ovarian cancer proliferation inhibition is mediated by PLAGL2 through mRNA stability regulation. In mechanism research, knockdown IGF2BP1 inhibits cell proliferation and is downstream gene PLAGL2 mRNA stability. Like IGF2BP1, unregulated PLAGL2 indicates a poor prognosis in ovarian cancer. Together this indicates that IGF2BP1 knockdown represses ovarian cancer cell proliferation largely by regulating target gene PLAGL2 mRNAs stability.







