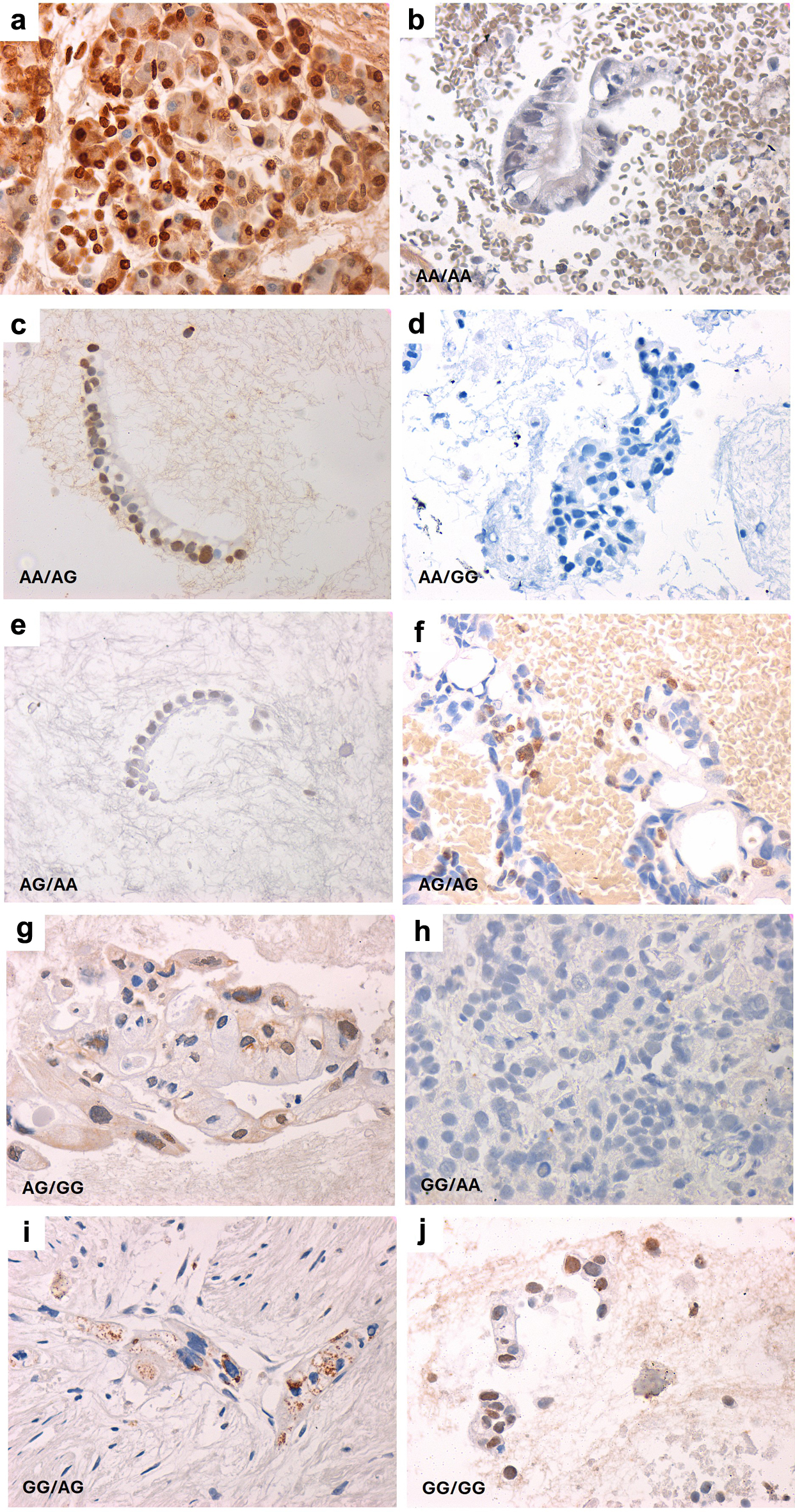
Figure 1. Representative images of VDR expression in pancreatic cancer according to VDR variants’ haplotype. VDR haplotypes are shown as rs2228570/rs731236. (a) Normal VDR expression in a healthy pancreatic specimen. (b-j) Representative images of pancreatic cancer cell block sections according to VDR haplotypes. All images were captured at × 400 magnification.