| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 13, Number 3, June 2022, pages 107-116
Promoter Gene Methylation Regulates Clooxygenase-2 Expression in Androgen-Dependent and Independent Prostate Cancer Cells
Chung-Yi Liua, b, c, Shih-Huan Sub, c, Tzu-Hsuan Changb, Ming-Li Hsiehb, c, Ying-Hsu Changa, b, c, Jacob See-Tong Pangb, c, d, e , Cheng-Keng Chuangb, c, d, e
aDepartment of Urology, New Taipei Municipal Tucheng Hospital, Chang Gung Memorial Hospital, New Taipei City 236017, Taiwan
bDivision of Urology, Department of Surgery, Chang Gung Memorial Hospital at Linkou, Taoyuan 33305, Taiwan
cDepartment of Medicine, College of Medicine, Chang Gung University, Taoyuan 33305, Taiwan
dGraduate Institute of Clinical Medical Science, College of Medicine, Chang Gung University, Taoyuan 33305, Taiwan
eCorresponding Author: Jacob See-Tong Pang and Cheng-Keng Chuang, Division of Urology, Department of Surgery, Chang Gung Memorial Hospital at Linkou, Guishan, Taoyuan 33305, Taiwanand
Manuscript submitted March 23, 2022, accepted June 4, 2022, published online June 22, 2022
Short title: Methylation of COX-2 in Prostate Cancer Cell
doi: https://doi.org/10.14740/wjon1478
| Abstract | ▴Top |
Background: Clooxygenase-2 (COX-2) expression is overexpressed in human prostate cancer, and aberrant methylation of the COX-2 promoter has also been elucidated. However, how the methylation of CpG islands at COX-2 regulates its expression in prostate cancer is still unclear. We will determine the methylated 5′ CpG island of the COX-2 gene and its role in the expression of COX-2 in prostate androgen-dependent and androgen-independent cancer cells, LNCaP and DU145.
Methods: We used western blotting and quantitative reverse transcription polymerase chain reaction (qRT-PCR) to confirm the COX-2 expression in prostate cancer cell lines, including LNCaP (androgen-dependent) and DU145 (androgen-independent) cells. To investigate whether the COX-2 expression was regulated by the methylation status of the 5′ CpG island, we treated LNCaP and DU145 cells with the DNA methylation inhibitor, 5-aza-2′-deoxycytidine, and determined COX-2 expression in the treated/untreated cells by western blotting and qRT-PCR. Subsequently, bisulfite sequencing was performed to study the methylation sites in the treated/untreated cells. The effects of 5-aza-2′-deoxycytidine to cell proliferation, cell migration and cell cycle process in DU145 and LNCaP cells were determined using Cell Counting Kit-8 (CCK-8) assay, transwell assay and flow cytometry, respectively.
Results: The results revealed that the expression of COX-2 in androgen-dependent LNCaP cells was 5.44-fold (in protein level) and 2.46-fold (in mRNA level) higher than that in androgen-independent DU145 cells. After 5-aza-2′-deoxycytidine treatment, COX-2 expression in DU145 cells was elevated significantly, but no change was found in LNCaP cells. The A and C regions of the COX-2 CpG island exhibited reduced methylation along with that an increased expression of COX-2 was noted in DU145 cell after 5-aza-2′-deoxycytidine treatment. Also, the treatment with 5-aza-2′-deoxycytidine inhibited cell proliferation, cell migration and influenced the cell cycle progression in both DU145 and LNCaP cells.
Conclusions: Our results reveal that androgen receptor (AR)-dependent/independent prostate cancer cell lines exhibit different regulation of methylation in COX-2 that regulate its expression. Additionally, 5-aza-2′-deoxycytidine treatment of DU145 and LNCaP cells inhibits their ability of tumor progression.
Keywords: Cyclooxygenase-2; Methylation; 5-aza-2′-deoxycytidine; Prostate cancer; Bisulfite sequencing
| Introduction | ▴Top |
Changes in the methylation status of CpG islands are frequently observed in numerous human cancers, including prostate cancer [1]. Aberrant methylation of CpG islands at glutathione S-transferase Pi 1 (GSTP1), adenomatous polyposis coli (APC), Ras association domain family 1 isoform A (RASSF1a), multidrug resistant 1 (MDR1), and cyclooxygenase-2 (COX-2; also known as prostaglandin-endoperoxide synthase (PTGS2)) genes have been discovered in most prostate cancer cell lines and tissues but not in their normal counterparts [2, 3]. Hypermethylation of CpG islands in the promoter region of the GSTP1 gene has been described as a highly specific and sensitive biomarker for prostate cancer [4].
It is often correlated to chromatin organization and alterations in gene expression by recruitment of histone deacetylases (HDACs) and methyl-CpG binding domain proteins (MBDs), which generally causes chromatin silencing [5, 6]. Alterations in DNA methylation in malignancies have been exploited for developing diagnostic tools for various cancers and can also be used to determine drug sensitivity [7]. DNA methyltransferase inhibitors (DNMTi), useful chemical tools, are widely used for hypomethylating the genomic DNA, that can investigate the roles of DNA methylation in biological processes [8]. DNMTi drugs, the nucleoside analogs 5-azacytidine (5-AZA-CR) and 5-aza-2′-deoxycytidine (5-AZA-dC), are assumed incorporated into replicating DNA, which results in the inhibition of DNA methyltransferases. 5-AZA-dC, a potent DNA methyltransferase 1 (DNMT1) inhibitor, incorporated into DNA, inhibits DNMT1 activity to induce loss of DNA methylation [9, 10]. Therefore, 5-AZA-dC was usually used to explore the correlation between demethylation of sites in specific genes and activation of the associated genes [8]. 5-AZA-CR and 5-AZA-dC have been approved by the Food and Drug Administration (FDA) for treating patients with hematological malignancies, including myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) [11, 12], and are also being tested as therapeutic options in multiple solid cancers [13, 14].
COX is an enzyme that converts arachidonic acid to prostaglandins (PGs). Two isozymes of COX found in humans, constitutive COX-1 and inducible COX-2, exhibit distinct functional differences, tissue distribution, and regulation of expression [15, 16]. COX-2 expression is induced by cytokines, growth factors, pro-inflammatory molecules, and tumor inducers [15]. The inflammatory mediator prostaglandin E2 (PGE2), which is catalyzed by COX-2, participates in several biological processes, including development, immune response, angiogenesis, and cancer progression [16]. Many factors influence COX-2 expression, such as GSTP1 and retinoic acid receptor β (RARβ) [17], which may be involved in the pathogenesis of cancer [18]. Various studies have revealed the upregulation of COX-2 in prostate cancer tissues and cell lines by using semi-quantitative reverse transcription-polymerase chain reaction (PCR), immunoblotting, and immunohistochemistry assays [18-20]. Aberrant methylation of the COX-2 promoter in numerous cancers, including prostate cancer, has also been reported [21-23]; however, how the methylation of CpG islands at COX-2 regulates its expression in prostate cancer is still unclear.
In this study, we will determine the methylated 5′ CpG island of the COX-2 gene and its role in the expression of COX-2 in prostate androgen-dependent and androgen-independent cancer cells.
| Materials and Methods | ▴Top |
Cell culture
Human prostate cancer cell lines, LNCaP (CLS Cat# 300265/p761_LNCaP, RRID:CVCL_0395) and DU145 (NCI-DTP Cat# DU-145, RRID:CVCL_0105), were purchased from the Bioresource Collection and Research Center (BCRC, Taiwan). LNCaP cells are androgen-sensitive human prostate adenocarcinoma cells with androgen receptor (AR) expression, which was established from a lymph node metastasis tissue. DU145 is an androgen-independent prostate cancer cell line, which lacks AR expression and was isolated from a brain metastasis prostate cancer patient. LNCaP cells were cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/mL penicillin, 100 µg/mL streptomycin (Gibco; Thermo Fisher Scientific, Inc.), and 2 mM L-glutamine (Gibco; Thermo Fisher Scientific, Inc.). On the other hand, DU145 cells were cultured in Eagle’s minimum essential medium (EMEM; ATCC, Manassas, VA, USA) containing 10% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM L-glutamine. All cells were maintained in a humidified atmosphere of 5% CO2 in air at 37 °C.
Treatment of 5-AZA-dC
Cells were seeded into 10 cm Petri dishes at a density of 5 × 105 cells/well. After seeding for 24 h, the cells were treated with 5-AZA-dC (5-AZA-dC; Millipore Sigma, USA) for 5 and 7 days and incubated at 37 °C in a humidified atmosphere with 5% CO2. Cells were collected and preserved at -80 °C for subsequent examination.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The cells were collected, and RNA was extracted using a Rneasy Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. RNA samples were quantified using NanoDrop® 2000 (Thermo Fisher Scientific Inc., Carlsbad, CA, USA) and were then stored at -80 °C for subsequent qRT-PCR analysis. The RNA was reverse transcribed into cDNA using the SuperScript™ First-Strand Synthesis SuperMix Kit (Invitrogen; Thermo Fisher Scientific, Inc.). The qRT-PCR of COX-2 was performed using iQ™ SYBR® Green SuperMix (Bio-Rad Laboratories Inc., USA) in a total 20 µL volume in a BioRad iCycler iQ™ Real-Time Detection System according to the manufacturer’s instructions. The primers used for the detection of COX-2, p53, myc, snail, and β-actin were listed in Table 1, and the sequences of primers were obtained from the sequences of qSTAR qPCR primers (Origene, USA). The qRT-PCR reactions were performed in triplicate, and β-actin was used as an internal control.
 Click to view | Table 1. The Sequences of Primers Used in This Study for qRT-PCR |
Western blot analysis
Cells were lysed on ice for 10 min using PRO-PREPTM protein extraction solution (iNtRON Biotechnology Inc., Korea). Proteins were quantified by the Bradford method using spectrophotometry (NanoDrop Technology Inc.; Thermo Fisher Scientific Inc., Carlsbad, CA, USA). Fifty micrograms of protein samples were separated using a 12% SDS-PAGE which were subsequently transferred to a polyvinylidene fluoride membrane (Merck Millipore, USA). Membranes were blocked with tris-buffered saline containing 0.05% Tween-20 (TBST) and 5% non-fat milk at room temperature for 1 h and then probed with the following primary antibodies: COX-2 (1:1,000; Cell Signaling Technology Cat# 12282, RRID:AB_2571729) and β-actin (1:5,000; Sigma-Aldrich Cat# A5316, RRID:AB_476743) for 2 h at room temperature. After washing with TBST buffer, the membranes were incubated with secondary antibodies (1:5,000; Millipore Cat# AP307P, RRID:AB_11212848; Millipore Cat# AP308P, RRID:AB_11215796, International, Inc.) for 1 h. Blots were visualized using an ECL system (Merck Millipore, USA) and analysed using the UVP ChemStudio Plus Touch (Analytik Jena US, Germany). ImageJ (ver. 1.53k, RRID:SCR_003070) was used to quantify the density of bands.
Bisulfite sequencing
Bisulfite sequencing of prostate cell lines was performed according to the method of research in gastric carcinoma cells [22]. The CpG island was divided into five regions (A-E). Genomic DNA (2 µg) obtained from each prostate cell line was treated with sodium bisulphite (Millipore Sigma, USA), and the COX-2 promoter region (A-E) was subsequently amplified using the primers listed in Table 2. PCR products were purified using the QIAquick® Gel Extraction Kit (Qiagen, Germany) and cloned into a TA cloning vector (Invitrogen, Carlsbad, CA). Sequences of individual plasmid clones were determined by Sanger sequencing.
 Click to view | Table 2. The Sequences of Primers Used in Bisulfite Sequencing |
Morphology and cell proliferation
The morphology of DU145 or LNCaP cells with/without 10 µM 5-AZA-dC treatment was examined and photographed by inverted microscope (ZEISS Axio Vert.A1, Germany). Cells (1 × 103) were seeded into 96-well plates (Corning, Corning Inc., NY, USA) with complete media containing 10 µM 5-AZA-dC or dimethylsulfoxide (DMSO) for 120 h, and the cell proliferation assay was examined by Cell Counting Kit-8 (CCK-8; Sigma-Aldrich; Merck KgaA, Darmstadt, Germany) according to the manufacturer’s instructions. Absorbance of each well was measured at 450 nm with an ultra-microplate reader (SpectraMax® ABS Plus, Molecular Devices, USA). The viability of the cells was measured in triplicate wells every time points for 120 h.
Cell migration assay
The cell migration assay was performed to add the cells directly to the Transwell® Permeable Support 8 µm PolyCarbonate Membrane in a 24-well plate (Corning). The cells at the bottom of the membrane were fixed in methanol and stained by Giemsa staining. Cells in at least four randomly selected microscopic fields (× 400) were counted. Each experiment was done in triplicate.
Analysis of cell cycle
DU145 cells (5 × 105) or LNCaP cells were seeded in six-well plates with complete media containing 10 µM 5-AZA-dC or DMSO for 72 h. Cells (1 × 106) were harvested and washed in ice cold phosphate-buffered saline (PBS) twice. Cells were fixed in 70% ethanol at -20 °C for at least 30 min, then washed with ice cold PBS once. Cells were resuspended in permeabilizing buffer (0.5% Triton X-100 and 200 µg/mL Rnase in PBS) and incubated at 37 °C for 1 h. Propidium iodide were added to samples (50 µg/mL) and incubated at room temperature for additional 10 min. The cell cycle status was analyzed using a flow cytometer (BD LSRFortessa™ Cell Analyzer, USA), and the cell cycle distribution was obtained from cell DNA content analysis by using FlowJo software (ver. 10.4.0, RRID:SCR_008520).
Statistical analysis
All assays were performed in triplicate, and results were represented as mean ± standard deviation (SD). Two-tailed unpaired Student’s t-test was used to compare two groups. The P < 0.05 was considered to indicate a statistical significance.
In our study, we only use prostate cell lines as study model, and there are no experiments included human/animal study. Our experiments in this study do not need Institutional Review Board Approval or ethical compliance.
| Results | ▴Top |
Higher expression of COX-2 in androgen-dependent LNCaP cells than that in androgen-independent DU145 cells
We used western blotting and qRT-PCR to confirm the expression of COX-2 in prostate normal/cancer cell lines, including androgen-dependent and androgen-independent cell lines. The results revealed that the expression of COX-2 in androgen-dependent LNCaP cells was 4.33-fold higher than that in androgen-independent DU145 cells (Fig. 1). To investigate whether the expression of COX-2 was regulated by the methylation status of the 5′ CpG island, we treated LNCaP and DU145 cells with the DNA methylation inhibitor, 5-AZA-dC (10 µM) and determined COX-2 expression in the treated cells by western blotting and qRT-PCR. The results showed that COX-2 expression in DU145 cells was elevated significantly but that in LNCaP cells were similar after 5-AZA-dC treatment (Fig. 2). Taken together, expression of COX-2 was low in DU145 cells, but expression of COX-2 was elevated after 5-AZA-dC treatment; expression of COX-2 was high in LNCaP cells, but expression of COX-2 was not induced after 5-AZA-dC treatment. This suggests that the expression of COX-2 in androgen-dependent and androgen-independent prostate cancer cells is regulated by different mechanisms.
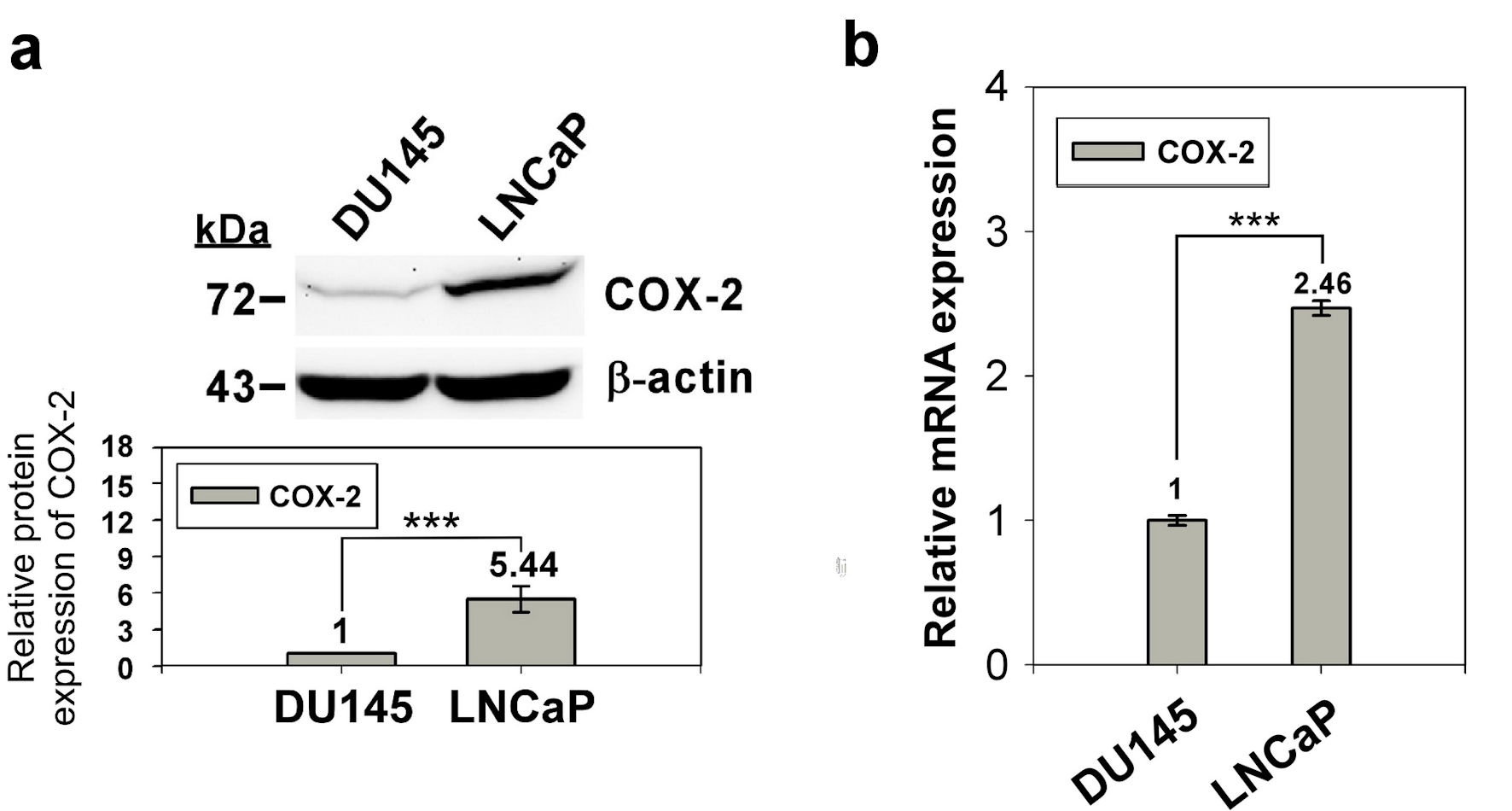 Click for large image | Figure 1. Expression of COX-2 in prostate normal/cancer cell lines. The protein expression of in DU145 and LNCaP cell was detect by western blotting (a) and the transcript expression was detected by qRT-PCR (b). ImageJ (version 1.53k) was used to quantify the density of bands in western blotting. The numbers under the bands indicate the relative expression, which normalized by the expression of β-actin. Experiments were performed in triplicate and repeated three times with similar results. All P values were determined by a two-tailed unpaired Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). COX-2: clooxygenase-2; qRT-PCR: quantitative reverse transcription polymerase chain reaction. |
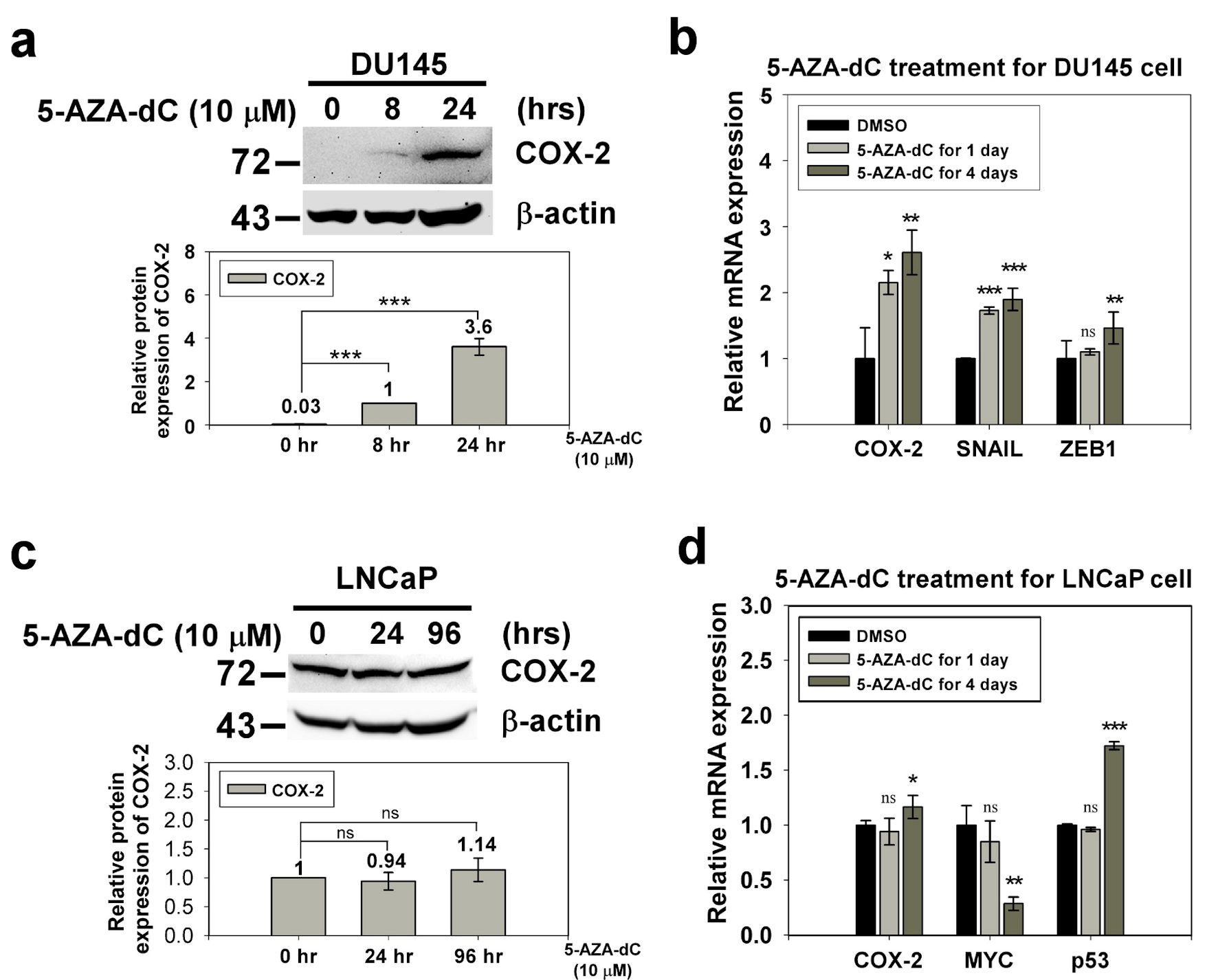 Click for large image | Figure 2. The expression of COX-2 in the DU145 and LNCaP cell with 5-AZA-dC treatment. DU145 and LNCaP cell were treated with 10 µM 5-AZA-dC for 1 and 4 days. The expression of COX-2 in treated cells were detected by western blotting (a, c) and qRT-PCR (b, d). SNAIL and ZEB1 were positive control of 5-AZA-dC treatment in DU145 cell. p53 and MYC were positive control and negative control of 5-AZA-dC treatment in LNCaP cell, respectively. ImageJ (version 1.53k) was used to quantify the density of bands in western blotting. The numbers under the bands indicate the relative expression, which was normalized by the expression of β-actin. Experiments were performed in triplicate and repeated three times with similar results. All P values were determined by a two-tailed unpaired Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). COX-2: clooxygenase-2; qRT-PCR: quantitative reverse transcription polymerase chain reaction; 5-AZA-dC: 5-aza-2′-deoxycytidine; DMSO: dimethylsulfoxide. |
Different methylation sites, in COX-2 CpG island, regulate COX-2 expression in androgen-dependent and androgen-independent prostate cancer cells
The aberrant methylation of the specific CpG island of the promoter is related to the expression of COX-2 in human gastric carcinoma cells [24]. To investigate the methylation of the 5′ CpG island of the COX-2 gene in androgen-dependent and androgen-independent prostate cancer cell, we treated the LNCaP and DU145 cells with 5-AZA-dC to inhibit their methylation. Bisulfite sequencing was performed to study the methylation sites in treated or untreated cells. The CpG island of COX-2 was divided into five regions (A to E), where A to C and E regions corresponded to the COX-2 promoter and D regions corresponded to exon 1 of the COX-2 gene (Fig. 3a) [23]. After 5-AZA-dC treatment of DU145 cells, the A and C regions of the COX-2 CpG island exhibited reduced methylation along with an increased expression of COX-2 were noted (Fig. 3b). Although 5-AZA-dC treatment of LNCaP cells revealed demethylation of the B and D regions of the COX-2 CpG island, the expression of COX-2 showed a slight increase (Fig. 3b). Hence, the expression of COX-2 was regulated not only by methylation of the 5′ CpG island of its promoter, but also by AR-related pathways in prostate cancer cell lines.
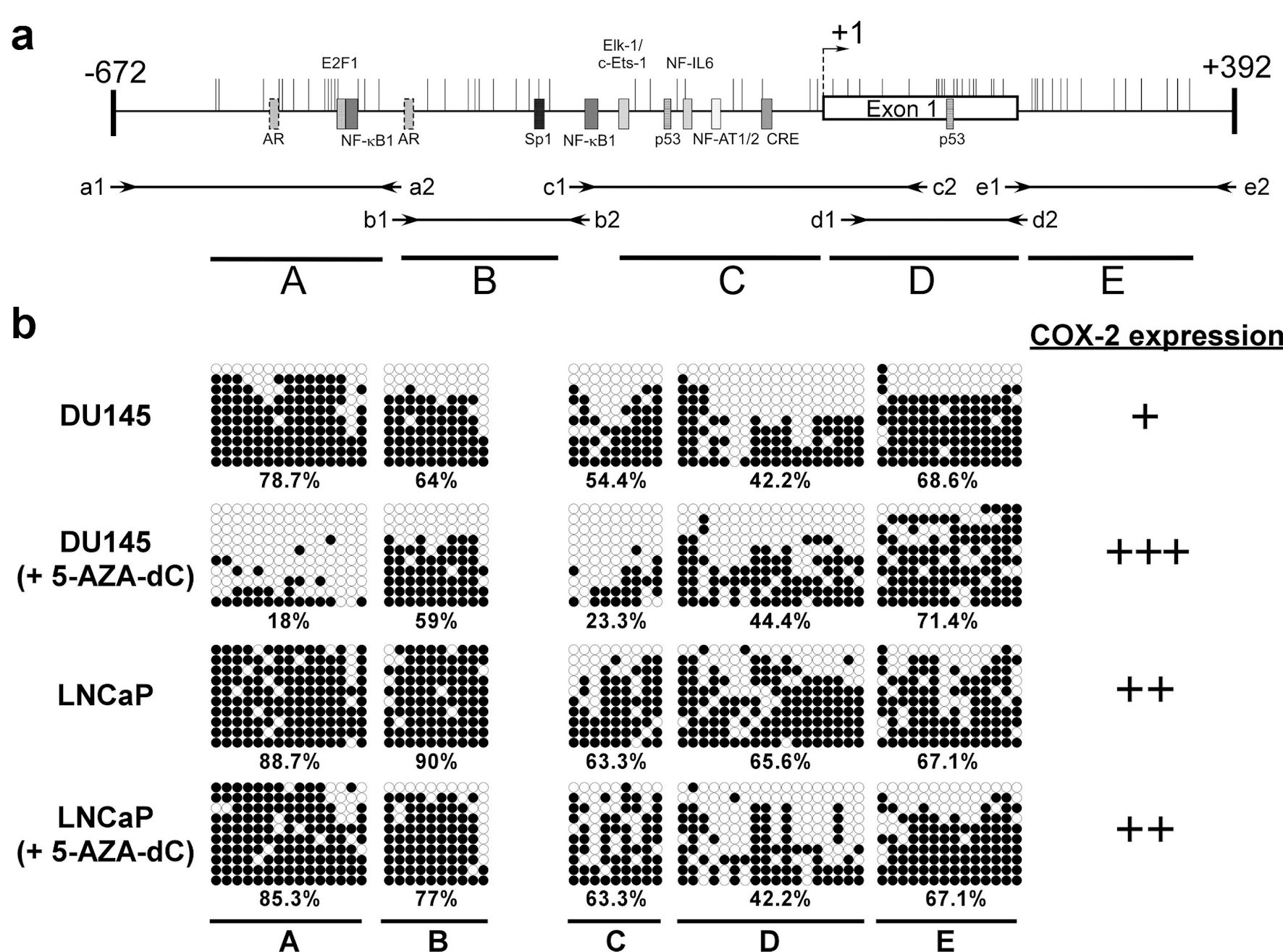 Click for large image | Figure 3. The methylation status of CpG islands at COX-2 promoter (-672 to +392) in prostate cancer cells. Schematic representation of the CpG islands (a). Horizontal bars indicate the location of CpG sites. CpG island was divided into five regions (A - E) defined by the specific primer sets used for PCR amplification (Table 2). The approximate locations and directions of the primers for each region are indicated by arrows denoted according to the segments to be amplified. The methylated sites of DU145 and LNCaP cell were detected by bisulfite sequencing (b). Each row of circles represents a single plasmid cloned and sequenced from PCR products generated from amplification of bisulfite-treated DNA. ○: unmethylated cytosines; •: methylated cytosines. The percentage of methylation rate is indicated below. COX-2: clooxygenase-2; PCR: polymerase chain reaction; 5-AZA-dC: 5-aza-2′-deoxycytidine. |
5-AZA-dC inhibits cell proliferation in DU145 and LNCaP cells
The cellular morphology of DU145 and LNCaP cells with or without 10 µM 5-AZA-dC treatment in indicated time points, 0 h, 48 h and 96 h, did not show any significant difference (Fig. 4a, b). To investigate the effects of 5-AZA-dC to the ability of proliferation in prostate cancer cells, the proliferation of the DU145 and LNCaP cells treated or untreated with 10 µM 5-AZA-dC for 120 h was examined by the CCK-8 assay. The results showed that the 5-AZA treatment inhibit the cell viability of DU145 and LNCaP cells significantly (Fig. 4c, d).
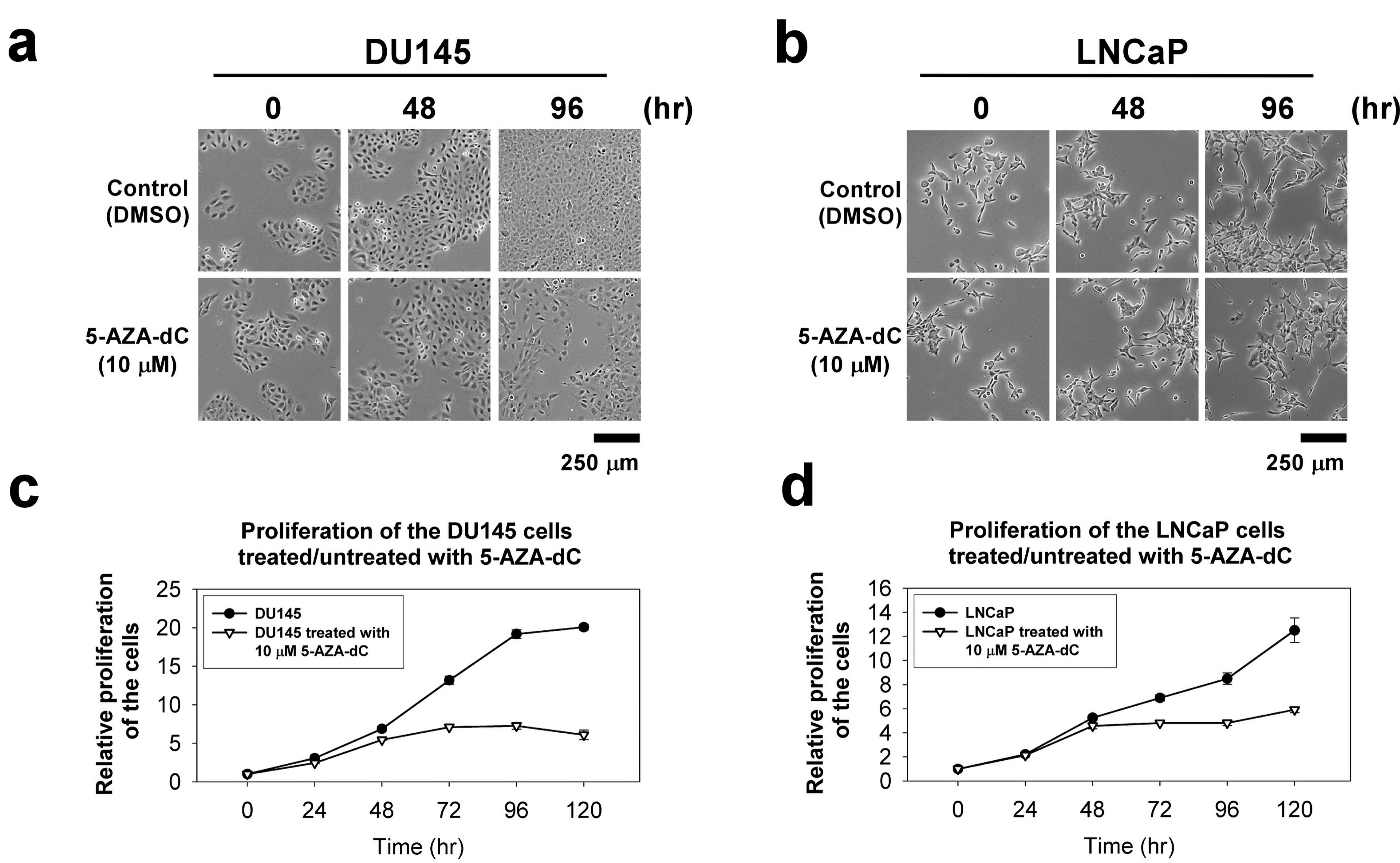 Click for large image | Figure 4. The cellular morphology and cell proliferation of DU145 and LNCaP cells treated/untreated with 10 µM 5-AZA-dC. Cells (1 × 103 cells) were seeded into a well of a standard 96-well plate and grown for 120 h in normal serum medium with/without 10 µM 5-AZA-dC. The cellular morphology of the cells was showed at indicated time point (a and b). The proliferation of the cells was measured by CCK-8 for 120 h (c and d). The data were presented as the mean ± SD of three independent experiments. CCK-8: Cell Counting Kit-8; 5-AZA-dC: 5-aza-2′-deoxycytidine; SD: standard deviation. |
5-AZA-dC inhibits cell migration in DU145 and LNCaP cells
The influence of 5-AZA-dC on the migration of DU145 and LNCaP cells was investigated by using a Transwell® system. Compared with the control DU145 cells, the cell migration of the DU145 cells with 5-AZA-dC treatment was significantly decreased (Fig. 5a up). In addition, compared with the control LNCaP cells, the cell migration of the LNCaP cells with 5-AZA-dC treatment was only slightly decreased (Fig. 5a down). The cell migration of the DU145 or LNCaP cells with or without 5-AZA-dC treatment was summarized and analyzed statistically (Fig. 5b).
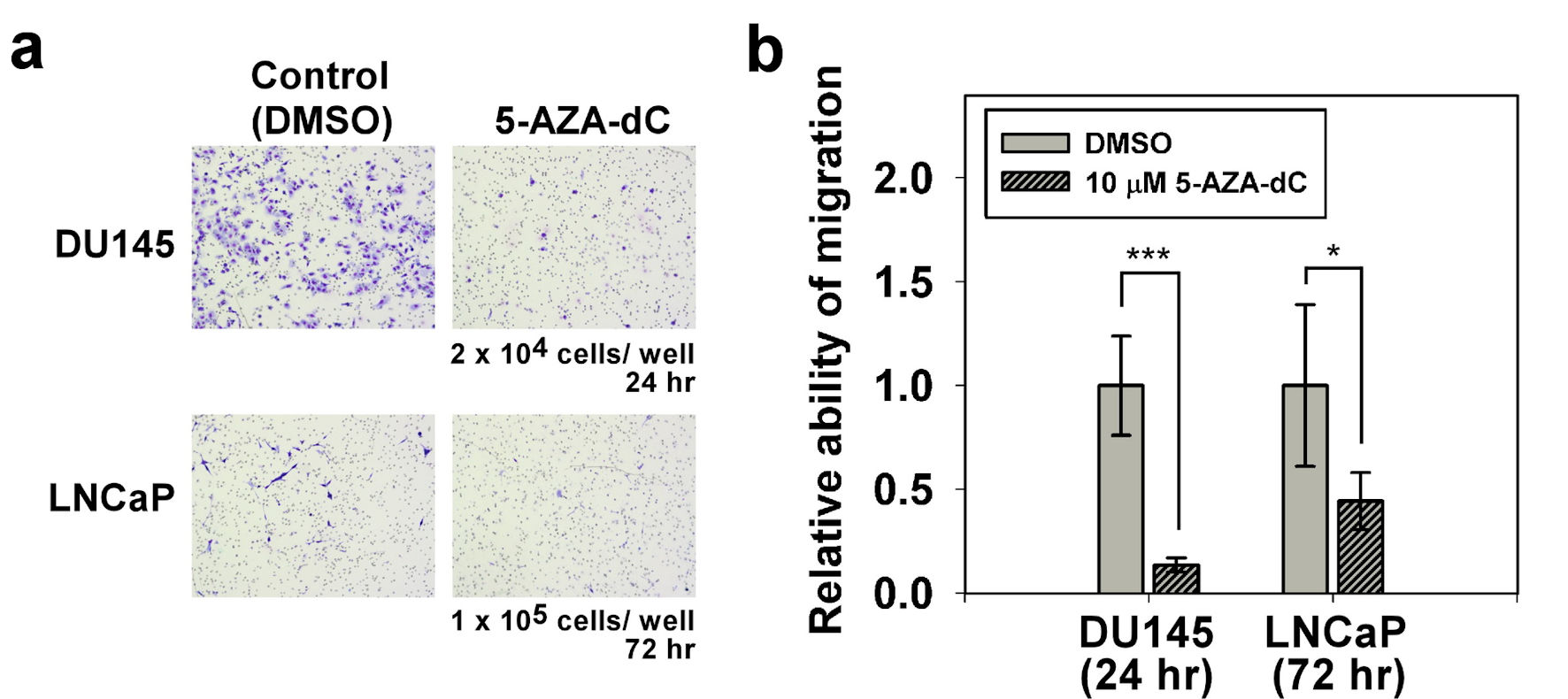 Click for large image | Figure 5. Migration assay of DU145 and LNCaP cells with/without 5-AZA-dC treatment. Transwell® assay was performed to assess the cell migration ability of DU145 and LNCaP cells with/without 10 µM 5-AZA-dC treatment. Cells were seeded into a transwell, and then cultured for indicated time point. Migrated cells were visualized by Giemsa staining. All P values were determined by a two-tailed unpaired Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). 5-AZA-dC: 5-aza-2′-deoxycytidine; DMSO: dimethylsulfoxide. |
5-AZA-dC affects cell cycle process in DU145 and LNCaP cells
To explore the effects of 5-AZA-dC on the cell cycle process, DU145 and LNCaP cells were treated with 10 µM of 5-AZA-dC for 72 h, and DNA of cells was stained with PI. Then, DNA contents in different cells were examined using flow cytometry assay, and cell cycle process of the cells was analyzed by FlowJo software. The results showed that treatment with 5-AZA-dC markedly increased the accumulation of cells at the Sub-G1 phase from 25.8% to 50.5% in LNCaP cells especially (Fig. 6a). Also, treatment with 5-AZA induced DU145 cells from G1 phase to G2/M phase, with G1 phase from 82.1% to 57.8% and G2/M phase from 6.41% to 26.5%, which blocked the cell cycle progression at G2/M phase (Fig. 6a). Our data revealed that the treatment of 5-AZA-dC affected cell cycle process in DU145 and LNCaP cells in different manners (Fig. 6).
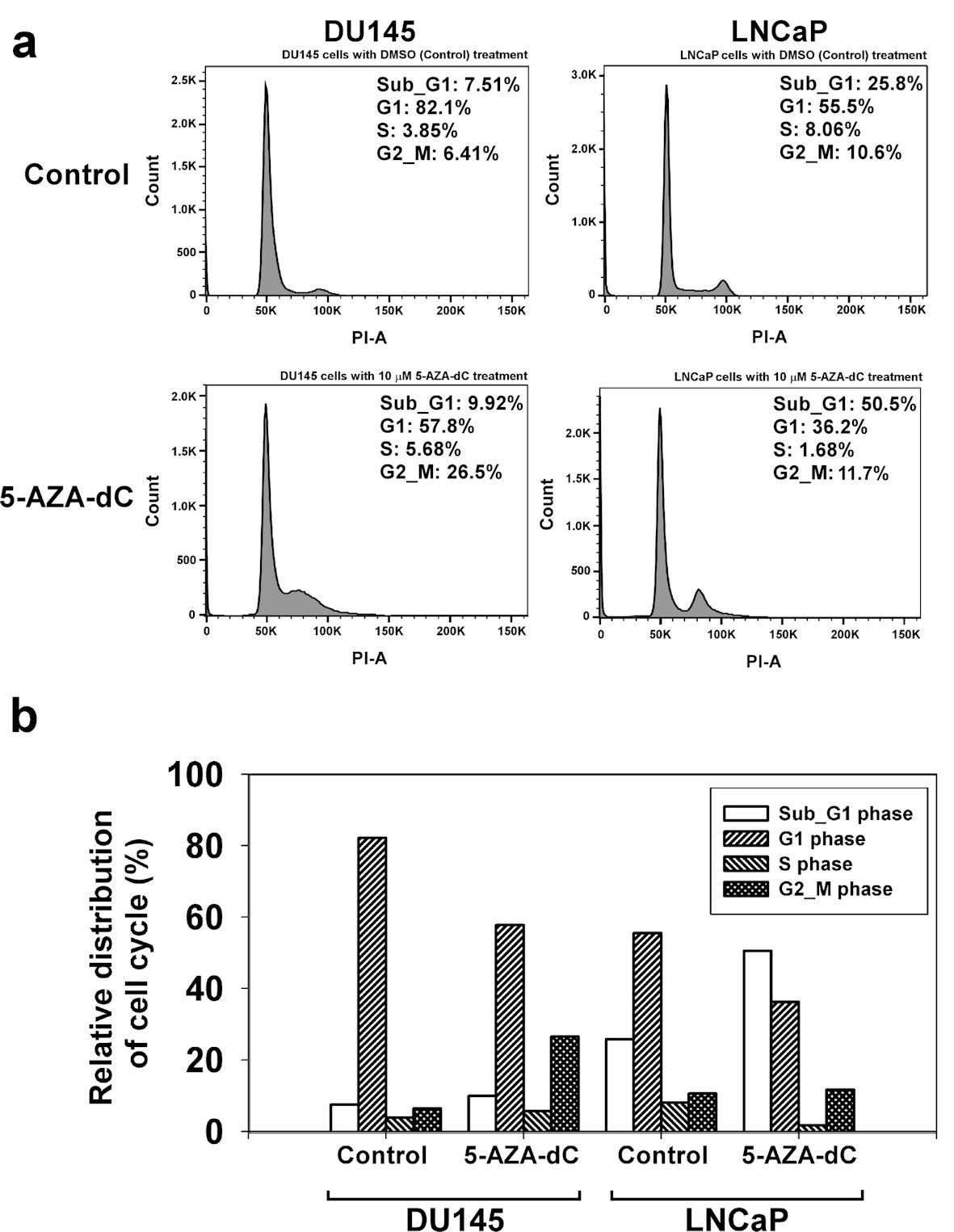 Click for large image | Figure 6. 5-AZA-dC affected the cell cycle process in DU145 and LNCaP cells. DU145 and LNCaP cells were treated with 10 µM 5-AZA-dC for 72 h. The cells were harvested, and the cell cycle status was analyzed by flow cytometry assay (a). The distribution of cell cycle in DU145 and LNCaP cells was summarized (b). 5-AZA-dC: 5-aza-2′-deoxycytidine; DMSO: dimethylsulfoxide. |
| Discussion | ▴Top |
In the present study, the results revealed the expression of COX-2 in androgen-dependent LNCaP cells higher than that in DU145 cells (androgen-independent). The expression of COX-2 did not correspond to the methylation status of the 5′ CpG island of the COX-2 promoter (Figs. 1, 3b), with LNCaP and DU145 cells exhibiting methylation percentages of 74.5% and 56.8%, respectively. However, it is noteworthy that different regions of the 5′ CpG island were methylated in these two cells. This indicates that the COX-2 expression is under the regulation of both the CpG island methylation status as well as other regulatory factors.
Till date, the regulation of COX-2 in cancer is not fully understood. Some studies have reported the involvement of transcription factors in the activation of COX-2 expression, including the nuclear factor-κB (NF-κB) [20, 25, 26], specificity protein-1 (Sp1) [27], cyclic AMP response element binding protein (CREB), erythroblast transformation-specific (Ets) family members Ets-1 and Elk-1 [28], nuclear factor of activated T cells (NF-AT1/2) [29] and p53 [30]. In addition, some methylated CpG sites of NF-IL6 and exon 1 are related to the expression of COX-2 [23, 31, 32]. Xu et al (2007) reported that epidermal growth factor receptor (EGFR) activation can transcriptionally activate COX-2 expression via p38 mitogen-activated protein kinase (p38-MAPK) and Sp1/Sp3 [27, 33]. Moreover, Jia et al (2008) reported that activation of EGFR induced expression through the PI-3K/AKT and/or p38-MAPK pathways in androgen-independent prostate cancer. Their results showed that both p38-MAPK and PI-3K/AKT pathways were involved in COX-2 upregulation in PC-3 cells, but only the p38-MAPK pathway was associated with COX-2 upregulation in DU145 cells [34]. These results revealed that the regulatory mechanisms of expression are complicated, and cell- and context-dependent [35]. In our results, LNCaP and DU145 cells treated with 5-AZA-dC, showed that COX-2 expression in the androgen-independent prostate cancer cell line DU145 was elevated significantly but in the androgen-dependent prostate cancer cell line LNCaP there was no change (Fig. 2). 5-AZA-dC treatment of DU145 cells reduced the methylation status of the A and C regions of the COX-2 CpG island and increased the expression of COX-2. Although 5-AZA-dC treatment of LNCaP cells revealed demethylation of the B and D regions of the COX-2 CpG island, along with a minor increase in the expression of COX-2 (Figs. 2, 3b). This is in alignment with other reports in the literature and suggests that COX-2 expression is under complex regulation of methylation status as well as other factors.
Song et al (2001) reported trans-activator binding sites, such as Sp1, NF-κB, NF-IL6, and CRE in the COX-2 gene promoter region (Fig. 3a) [22]. Other cis-acting elements are known to mediate cytokine-mediated promoter activity in the COX-2 gene, and most of them are in regions A and C of the CpG island of COX-2 promoter. Furthermore, there were two putative AR binding sites in region A of the COX-2 promoter. Taken together, we hypothesize that the methylation status of regions A and C is related to COX-2 expression in the prostate cancer cells. Toyota et al (2000) and Kikuchi et al (2002) reported that COX-2 expression was repressed by aberrant methylation of the exon 1 coding region of the COX-2 gene [23, 32]. Exon 1 was involved in region D of the COX-2 CpG island as per our study. However, the demethylation of regions B and D in the COX-2 gene in LNCaP did not restore the expression of COX-2 (Figs. 2, 3b). Hence, the mechanism by which different methylation regions of COX-2 CpG islands regulate the expression of COX-2 in prostate cancer should be further explored.
Our data revealed that the treatment of 5-AZA-dC inhibited the cell proliferation and ability of migration (Figs. 4, 5) and affected cell cycle process in the DU145 and LNCaP cells (Fig. 6). These results were consistent with the findings of Cheng et al [36]. We will investigate the functions of DNMT inhibitor in prostate cancer by using in vivo models in the future.
Conclusions
The expression of COX-2 was higher in androgen-dependent prostate cancer cells than in androgen-independent prostate cancer cells. Furthermore, the methylation of COX-2 CpG island in androgen-dependent prostate cancer cells may involve other methyl-CpG-binding proteins and factors in COX-2 gene regulation. Our results reveal that AR-dependent/independent prostate cancer cell lines exhibit different regulation of methylation in genes that regulate gene expression. In addition, the treatment of 5-AZA-dC inhibited the cell proliferation and cell migration and affected cell cycle process in the prostate cancer cells. Therefore, the role of methylation and its relationship with prostate cancer should be explored in more details in the future, to obtain new insights into our understanding of the pathogenesis of prostate cancer thereby facilitating the development of novel therapeutic treatments.
Acknowledgments
We wish to thank Tzu-Kai Wang and Ya-Ping Liu for their technical assistance.
Financial Disclosure
This study was supported by grants from Chang Gung Memorial Hospital of Taiwan (CORPG3H0051).
Conflict of Interest
The authors declare they have no conflict of interest to declare.
Informed Consent
Not applicable.
Author Contributions
Conception and design: CK Chuang and Jacob ST Pang. Administrative support: ML Hsieh. Provision of study materials: CY Liu. Collection and assembly of data: TH Chang. Data analysis and interpretation: SH Su and YH Chang. Manuscript writing and final approval of manuscript: all authors.
Data Availability
The dataset supporting the conclusions of this article is included within this article and is available from the corresponding author upon request.
Abbreviations
COX-2: clooxygenase-2; 5-AZA-CR: 5-azacytidine; 5-AZA-dC: 5-aza-2′-deoxycytidine; AR: androgen receptor
| References | ▴Top |
- Ilango S, Paital B, Jayachandran P, Padma PR, Nirmaladevi R. Epigenetic alterations in cancer. Front Biosci (Landmark Ed). 2020;25(6):1058-1109.
doi pubmed - Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64(6):1975-1986.
doi pubmed - Perry AS, Foley R, Woodson K, Lawler M. The emerging roles of DNA methylation in the clinical management of prostate cancer. Endocr Relat Cancer. 2006;13(2):357-377.
doi pubmed - Chuang CK, Chu DC, Tzou RD, Liou SI, Chia JH, Sun CF. Hypermethylation of the CpG islands in the promoter region flanking GSTP1 gene is a potential plasma DNA biomarker for detecting prostate carcinoma. Cancer Detect Prev. 2007;31(1):59-63.
doi pubmed - Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549-565.
doi pubmed - Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278(7):4806-4812.
doi pubmed - Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med (Zagreb). 2015;25(2):161-176.
doi pubmed - Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483-5495.
doi pubmed - Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457-463.
doi pubmed - Qiu X, Hother C, Ralfkiaer UM, Sogaard A, Lu Q, Workman CT, Liang G, et al. Equitoxic doses of 5-azacytidine and 5-aza-2′deoxycytidine induce diverse immediate and overlapping heritable changes in the transcriptome. PLoS One. 2010;5(9):e12994.
doi pubmed - Orta ML, Calderon-Montano JM, Dominguez I, Pastor N, Burgos-Moron E, Lopez-Lazaro M, Cortes F, et al. 5-Aza-2′-deoxycytidine causes replication lesions that require Fanconi anemia-dependent homologous recombination for repair. Nucleic Acids Res. 2013;41(11):5827-5836.
doi pubmed - Bohl SR, Bullinger L, Rucker FG. Epigenetic therapy: azacytidine and decitabine in acute myeloid leukemia. Expert Rev Hematol. 2018;11(5):361-371.
doi pubmed - Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15(12):3938-3946.
doi pubmed - Giri AK, Aittokallio T. DNMT Inhibitors Increase Methylation in the Cancer Genome. Front Pharmacol. 2019;10:385.
doi pubmed - Griswold DE, Adams JL. Constitutive cyclooxygenase (COX-1) and inducible cyclooxygenase (COX-2): rationale for selective inhibition and progress to date. Med Res Rev. 1996;16(2):181-206.
doi - Stasinopoulos I, Shah T, Penet MF, Krishnamachary B, Bhujwalla ZM. COX-2 in cancer: Gordian knot or Achilles heel? Front Pharmacol. 2013;4:34.
doi pubmed - Gupta S, Srivastava M, Ahmad N, Bostwick DG, Mukhtar H. Over-expression of cyclooxygenase-2 in human prostate adenocarcinoma. Prostate. 2000;42(1):73-78.
doi - Hussain T, Gupta S, Mukhtar H. Cyclooxygenase-2 and prostate carcinogenesis. Cancer Lett. 2003;191(2):125-135.
doi - Kirschenbaum A, Liu X, Yao S, Levine AC. The role of cyclooxygenase-2 in prostate cancer. Urology. 2001;58(2 Suppl 1):127-131.
doi - Garg R, Blando JM, Perez CJ, Lal P, Feldman MD, Smyth EM, Ricciotti E, et al. COX-2 mediates pro-tumorigenic effects of PKCepsilon in prostate cancer. Oncogene. 2018;37(34):4735-4749.
doi pubmed - Ma X, Yang Q, Wilson KT, Kundu N, Meltzer SJ, Fulton AM. Promoter methylation regulates cyclooxygenase expression in breast cancer. Breast Cancer Res. 2004;6(4):R316-321.
doi pubmed - Song SH, Jong HS, Choi HH, Inoue H, Tanabe T, Kim NK, Bang YJ. Transcriptional silencing of Cyclooxygenase-2 by hyper-methylation of the 5′ CpG island in human gastric carcinoma cells. Cancer Res. 2001;61(11):4628-4635.
- Toyota M, Shen L, Ohe-Toyota M, Hamilton SR, Sinicrope FA, Issa JP. Aberrant methylation of the Cyclooxygenase 2 CpG island in colorectal tumors. Cancer Res. 2000;60(15):4044-4048.
- Hur K, Song SH, Lee HS, Ho Kim W, Bang YJ, Yang HK. Aberrant methylation of the specific CpG island portion regulates cyclooxygenase-2 gene expression in human gastric carcinomas. Biochem Biophys Res Commun. 2003;310(3):844-851.
doi pubmed - Lim JW, Kim H, Kim KH. Nuclear factor-kappaB regulates cyclooxygenase-2 expression and cell proliferation in human gastric cancer cells. Lab Invest. 2001;81(3):349-360.
doi pubmed - Cai Y, Lee YF, Li G, Liu S, Bao BY, Huang J, Hsu CL, et al. A new prostate cancer therapeutic approach: combination of androgen ablation with COX-2 inhibitor. Int J Cancer. 2008;123(1):195-201.
doi pubmed - Xu K, Shu HK. EGFR activation results in enhanced cyclooxygenase-2 expression through p38 mitogen-activated protein kinase-dependent activation of the Sp1/Sp3 transcription factors in human gliomas. Cancer Res. 2007;67(13):6121-6129.
doi pubmed - Zhang X, Zhang J, Yang X, Han X. Several transcription factors regulate COX-2 gene expression in pancreatic beta-cells. Mol Biol Rep. 2007;34(3):199-206.
doi pubmed - Flockhart RJ, Diffey BL, Farr PM, Lloyd J, Reynolds NJ. NFAT regulates induction of COX-2 and apoptosis of keratinocytes in response to ultraviolet radiation exposure. FASEB J. 2008;22(12):4218-4227.
doi pubmed - Corcoran CA, He Q, Huang Y, Sheikh MS. Cyclooxygenase-2 interacts with p53 and interferes with p53-dependent transcription and apoptosis. Oncogene. 2005;24(9):1634-1640.
doi pubmed - Wang D, Chen Q, Zhang C, Ren F, Li T. DNA hypomethylation of the COX-2 gene promoter is associated with up-regulation of its mRNA expression in eutopic endometrium of endometriosis. Eur J Med Res. 2012;17:12.
doi pubmed - Kikuchi T, Itoh F, Toyota M, Suzuki H, Yamamoto H, Fujita M, Hosokawa M, et al. Aberrant methylation and histone deacetylation of cyclooxygenase 2 in gastric cancer. Int J Cancer. 2002;97(3):272-277.
doi pubmed - Hu H, Han T, Zhuo M, Wu LL, Yuan C, Wu L, Lei W, et al. Elevated COX-2 Expression Promotes Angiogenesis Through EGFR/p38-MAPK/Sp1-Dependent Signalling in Pancreatic Cancer. Sci Rep. 2017;7(1):470.
doi pubmed - Jia RP, Xu LW, Su Q, Zhao JH, Li WC, Wang F, Xu Z. Cyclooxygenase-2 expression is dependent upon epidermal growth factor receptor expression or activation in androgen independent prostate cancer. Asian J Androl. 2008;10(5):758-764.
doi pubmed - Modi PK, Faiena I, Kim IY. Chapter 3 - androgen receptor. In: Mydlo JH, Godec CJ, eds. Prostate cancer (Second Edition). Academic Press. 2016:21-28.
doi pubmed - Cheng H, Tang S, Lian X, Meng H, Gu X, Jiang J, Li X. The differential antitumor activity of 5-Aza-2′-deoxycytidine in prostate cancer DU145, 22RV1, and LNCaP Cells. J Cancer. 2021;12(18):5593-5604.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


