| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 14, Number 1, February 2023, pages 51-59
Uses of Vascular Endothelial Growth Factor C as a Lung Adenocarcinoma Prognostic Biomarker
Shi Chena, c, Ting Yu Pana, c, Xiao Wua, Tian Lib, Yu Weia, Hai Lang Hea, Xian Mei Zhoua, Qian Wanga, d, Ji Ping Zhua, d
aDepartment of Respiratory Medicine, Jiangsu Province Hospital of Chinese Medicine, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, Jiangsu Province, China
bDepartment of Respiratory Medicine of the Affiliated Brain Hospital of Nanjing Medical University, Nanjing, Jiangsu 210029, China
cThese authors contributed equally to this article.
dCorresponding Author: Qian Wang and Ji Ping Zhu, Department of Respiratory Medicine, Jiangsu Province Hospital of Chinese Medicine, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, Jiangsu Province, Chinaand
Manuscript submitted July 30, 2022, accepted December 16, 2022, published online February 26, 2023
Short title: VEGF-C in LUAD
doi: https://doi.org/10.14740/wjon1520
| Abstract | ▴Top |
Background: Lung adenocarcinoma (LUAD) is the most common type of lung cancer and a leading cause of death worldwide. Vascular endothelial growth factor C (VEGF-C) has been identified as a prognosis prediction marker for LUAD. However, VEGF-C protein expression does not appear to significantly relate to LUAD patient survival in several studies.
Methods: We carried out a bioinformatic analysis to review the effect of VEGF-C mRNA expression on LUAD patient outcomes. GEPIA, UALCAN, TCGAportal, OncoLnc, LCE, GeneMANIA, Metascape, ImmuCellAI, and GSCA online databases were utilized. The expression levels of VEGF-C mRNA between normal tissue and LUAD tissue, overall survival (OS) analysis, function analysis, tumor microenvironment and drug sensitivity were conducted in the current study.
Results: We found that the expression level of VEGF-C mRNA was significantly lower in LUAD than normal tissue. Low expression of VEGF-C mRNA was also associated with better OS. VEGF-C expression was correlated with both NF1 and TP53 mutation status. No relationship was observed between VEGF-C and Tr1 or CD4 T-cell infiltrate scores. Additionally, VEGF-C was associated with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance. The sensitivity of 5-fluorouracil was positively correlated with VEGF-C, and the sensitivity of TGX221 was negatively correlated with VEGF-C. The activity of BI-2536 and BRD-A94377914 was positively correlated with VEGF-C.
Conclusion: Novel LUAD prognostic biomarkers such as VEGF-C mRNA may aid diagnosis and treatment, and may help identify optimal LUAD populations for therapeutic treatments.
Keywords: Lung adenocarcinoma; VEGF-C; Survival; mRNA; P53
| Introduction | ▴Top |
Lung adenocarcinoma (LUAD) is the most common type of lung cancer, comprising around 40% of all lung cancer cases, and it is a leading cause of death worldwide [1, 2]. Most LUAD patients are unfortunately diagnosed at an advanced stage and consequently miss the best treatment time. Conventional LUAD treatments involving surgical resection, radiotherapy, and chemotherapy often have a poor prognosis, with a 5-year survival rate ranging between 5% and 16% [3]. Newer treatments such as molecular targeted therapy and immunotherapy, however, generate long-lasting responses and improved survival rates [4].
Predicting LUAD prognosis is needed to develop a reasonable follow-up treatment plan and to calibrate treatment. Vascular endothelial growth factor C (VEGF-C) has been identified as a prognosis prediction marker for LUAD in several studies [5, 6]. However, VEGF-C protein expression does not appear to significantly relate to LUAD patient survival. Yang et al found that VEGF-C mRNA expression correlated with survival of the patients with lung cancer [7]. The biological functions of VEGF-C may be analyzed as a potential prognosis factor in LUAD.
Newly developed sequencing technologies can detect large-scale tumor-related genes. We therefore carried out a bioinformatic analysis to review the effect of VEGF-C mRNA expression on survival in patients with LUAD. We used several online databases including GEPIA, UALCAN, TCGAportal, OncoLnc, LCE, GeneMANIA, Metascape, ImmuCellAI, and GSCA to explore the expression level of VEGF-C mRNA between normal tissue and LUAD tissue. Overall survival (OS) analysis, function analysis, tumor microenvironment and drug sensitivity were also conducted in the current study.
| Materials and Methods | ▴Top |
Expression level of VEGF-C mRNA in LUAD
GEPIA [8], Ualcan [9] and TCGAportal [10] were applied to evaluate the expression of VEGF-C in LUAD. Forms of VEGF-C mRNA expression in subgroups of LUAD were subdivided based on stage and gene mutation status.
OS analysis
GEPIA [8], Ualcan [9], OncoLnc [11], and TCGAportal [10] were used to evaluate the impact of expression levels of VEGF-C mRNA on OS in LUAD patients. OS curves were plotted using Kaplan-Meier and log-rank tests. LCE was used to combine multiple datasets to evaluate the prognostic precision of VEGF-C [12]. Different cut-off values were used to divide the high and low expression levels of VEGF-C mRNA. Samples were considered “high expression” when samples with gene expression values equal to or more than the third quartile value; otherwise, samples were considered “low/medium expression” (Fig. 1a) [13]. The median of expression levels of VEGF-C mRNA is the cut-off value for dividing high and low expression groups (Fig. 1b-d) [14].
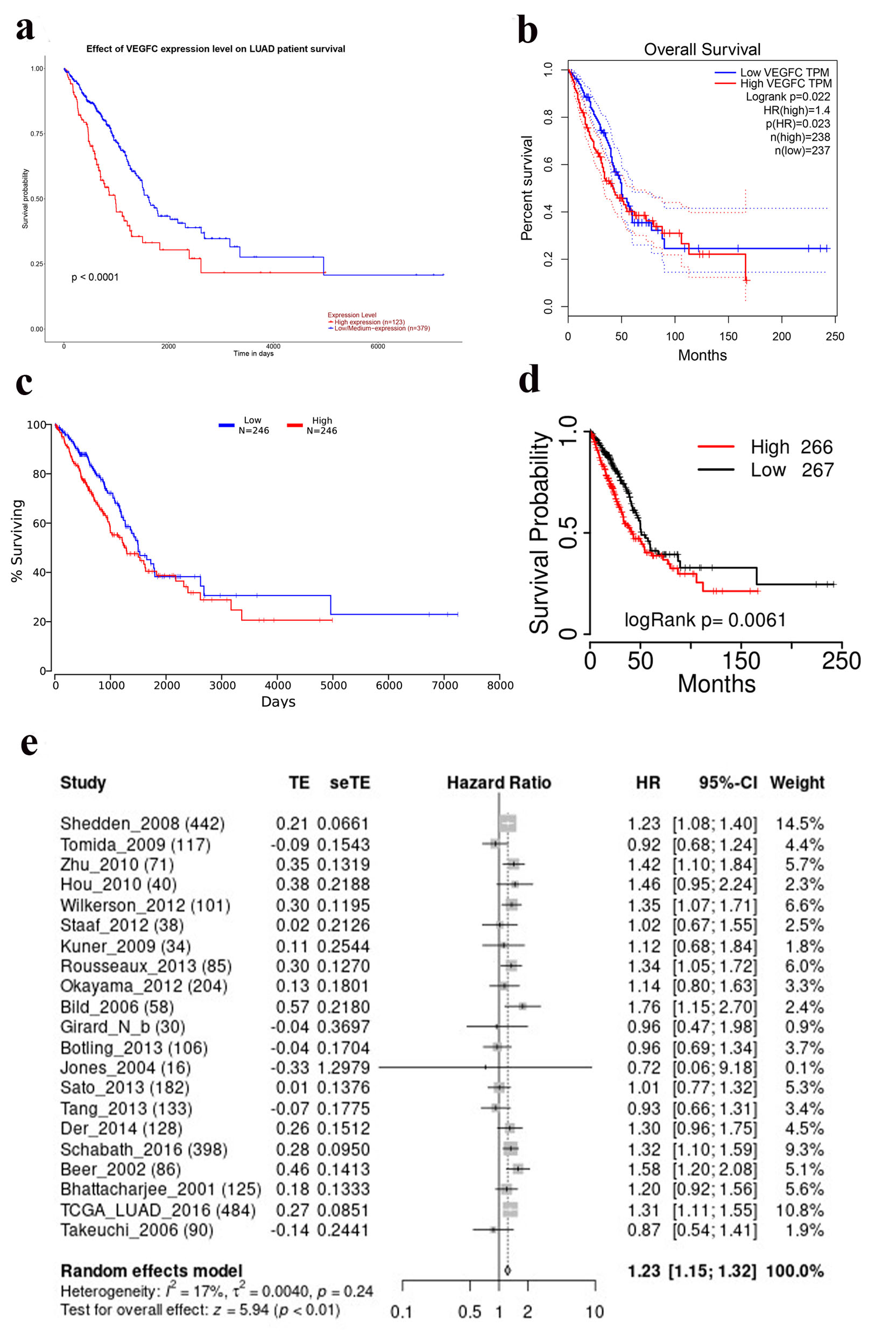 Click for large image | Figure 1. Overall survival curves of VEGF-C in LUAD from different database (a) Ualcan. (b) GEPIA. (c) OncoLnc. (d) TCGA portal. (e) Results of meta-analysis from LCE. LUAD: lung adenocarcinoma; VEGF-C: vascular endothelial growth factor C. |
Function analysis of VEGF-C
GeneMANIA was used to generate hypotheses about gene function of VEGF-C [15]. Gene Ontology (GO) categorization and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted to understand the functions of VEGF-C by using Metascape [16].
Tumor microenvironment and VEGF-C
ImmuCellAI was used to estimate the impact of 24 immune cells on the OS of LUAD patients [17]. GSCA was used to detect the relationship between VEGF-C and immune microenvironment in LUAD [18].
Prediction of drug sensitivity based on VEGF-C
GSCA was used to detect correlation between VEGF-C expression and small molecule drug sensitivity of GDSC and CTRP [18].
| Results | ▴Top |
Expression level of VEGF-C mRNA in LUAD
We compared the expression level of VEGF-C mRNA between 59 normal lung tissue and 515 LUAD tissue, 483 normal lung tissue and 347 LUAD tissue, respectively. The expression level of VEGF-C mRNA was significantly higher in normal lung tissue than in LUAD tissue (Fig. 2a, b). Patients with LUAD patients were divided into four stages. Different stages of LUAD patients had significantly different levels of VEGF-C mRNA expression (Fig. 2c). Of 111 normal lung tissue and 111 LUAD tissue, no significant difference was observed in VEGF-C protein levels (Fig. 2d). Among genes of TP53, KRAS, KEAP1, STK11, EGFR, NF1 and BRAF, only the mutation status of TP53 and NF1 was correlated with the expression level of VEGF-C (Fig. 2e).
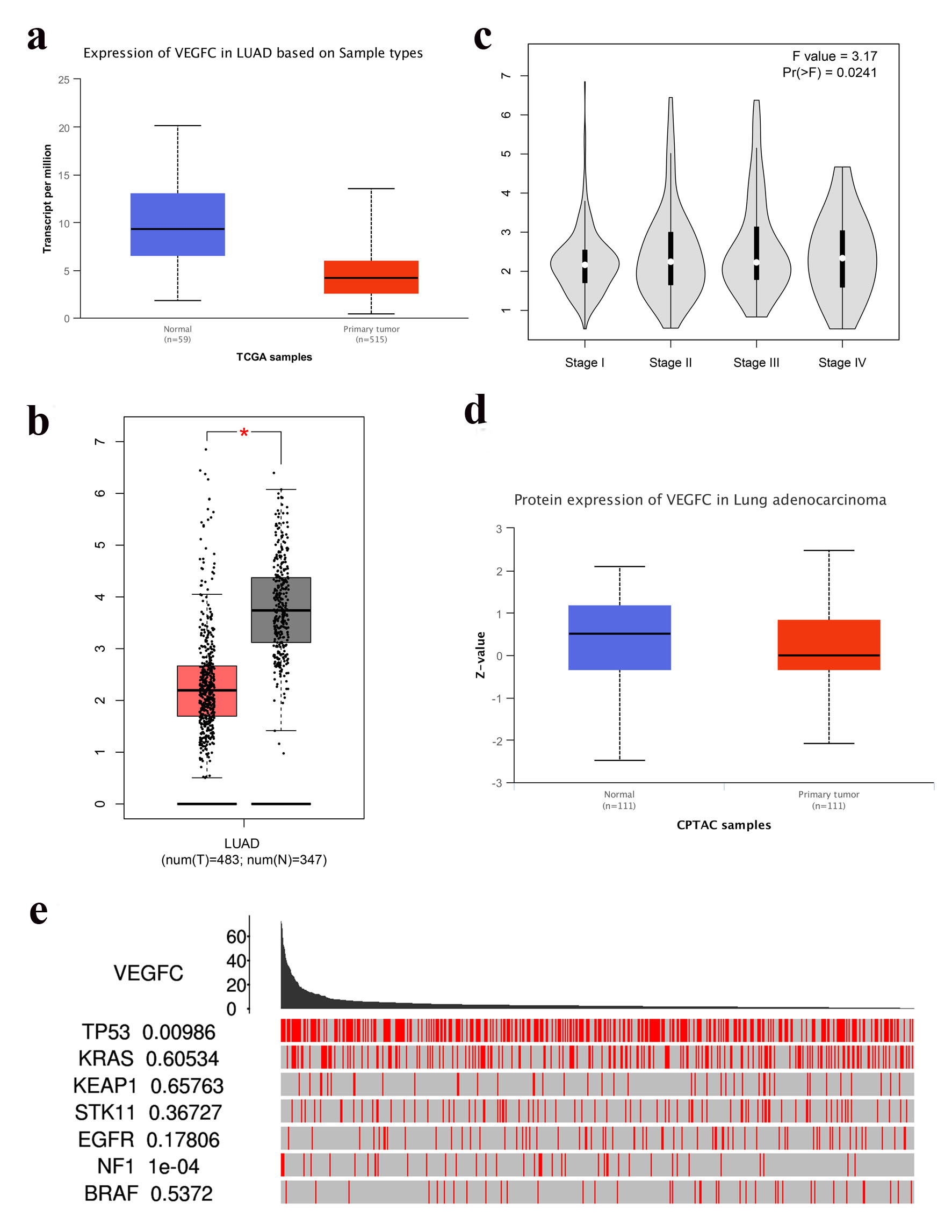 Click for large image | Figure 2. (a) VEGF-C mRNA expression in LUAD (Ualcan, P = 7.41 × 10-3). (b) VEGF-C mRNA expression in LUAD (GEPIA, P < 0.05). (c) VEGF-C mRNA expression in LUAD based on stage (GEPIA, P = 0.0241). (d) VEGF-C protein expression in LUAD (Ualcan, P = 0.195). (e) VEGF-C mRNA expression between driver mutated (red) and not-mutated (gray) samples (TCGA portals). LUAD: lung adenocarcinoma; VEGF-C: vascular endothelial growth factor C. |
VEGF-C mRNA expression predicts OS
OS analyses were conducted on 123 samples with high expression levels of VEGF-C mRNA and 379 samples with low/medium levels of VEGF-C mRNA, 238 samples with high expression levels of VEGF-C mRNA and 237 samples with low levels of VEGF-C mRNA, 246 samples with high expression levels of VEGF-C mRNA and 246 samples with low levels of VEGF-C mRNA, 266 samples with high expression levels of VEGF-C mRNA and 267 samples with low levels of VEGF-C mRNA, respectively. OS of LUAD with low VEGF-C mRNA expression was significantly increased compared to LUAD with high VEGF-C mRNA expression (Fig. 1a-d). Twenty-one studies were included in meta-analysis. Figure 1e shows that high VEGF-C mRNA expression was associated with inferior LUAD OS (hazard ratio (HR) = 1.23, 95% confidence interval (CI), 1.15 - 1.32, Fig. 1e). These results revealed that VEGF-C mRNA is a valuable prognostic biomarker in LUAD.
Functional analysis of VEGF-C
GeneMANIA database analysis showed that VEGF-C interacted with genes including FLT4, VEGFD, KDR, NRP2, EPAS1, VEGFA, FLT1, PDGFRA, PDGFA, VEGFB, PGF, LOXL2, IGF1R, ITGA9, PDGFB, NRP1, HHEX, ERBB2, NID2, and MET (Fig. 3a). The GO analysis showed that the differentially expressed genes were mainly related to VEGF, platelet-derived growth factor and endothelial cell (Fig. 3b). In the KEGG signal pathway, LUAD mostly involved in the focal adhesion signaling pathway, pathways in EGFR tyrosine kinase inhibitor (TKI) resistance and transcriptional misregulation in cancer signaling pathway (Fig. 3c).
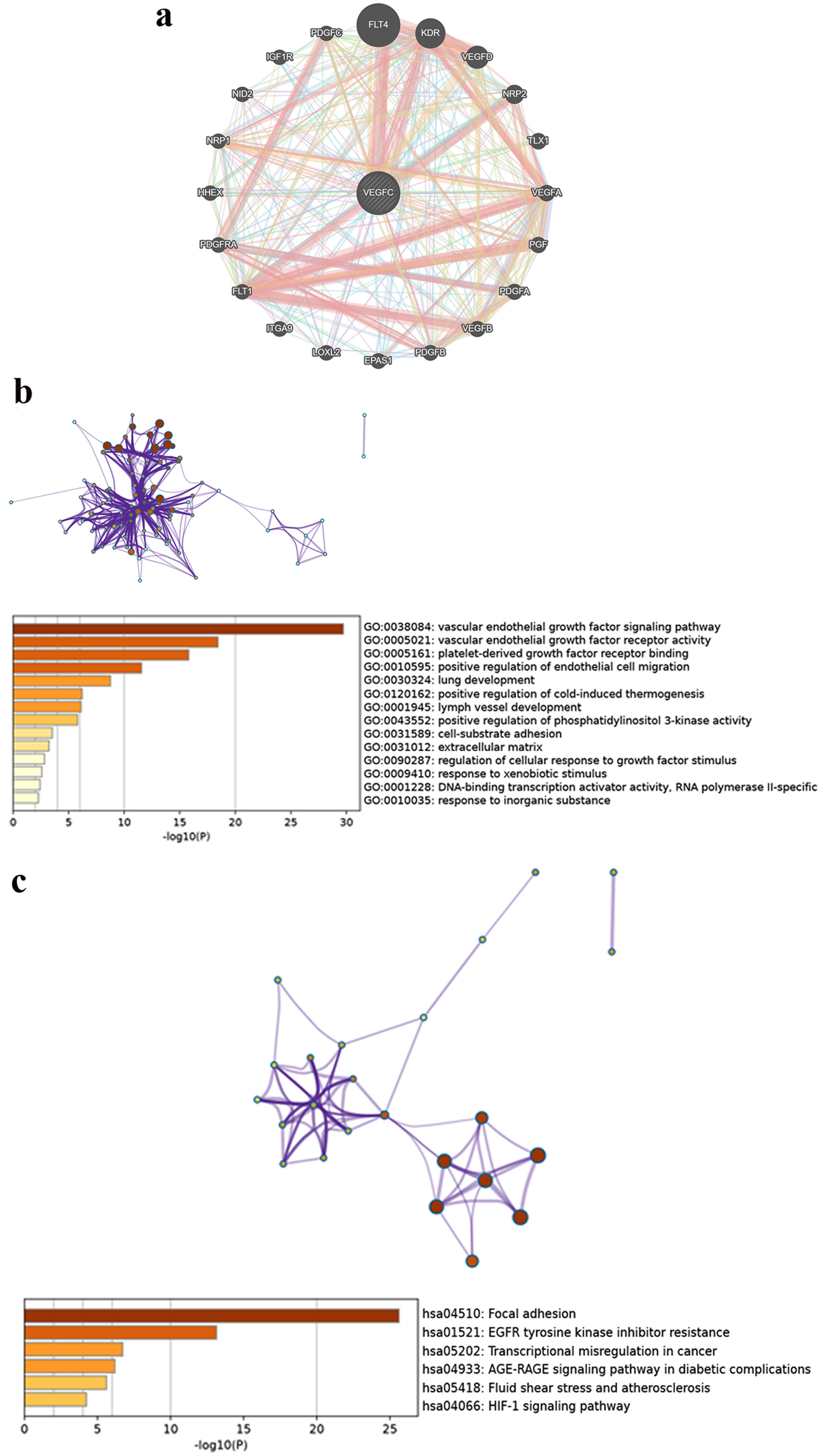 Click for large image | Figure 3. (a) A composite gene-gene functional interaction network. (b) GO of VEGF-C in LUAD. (c) KEGG pathway of VEGF-C in LUAD. LUAD: lung adenocarcinoma; VEGF-C: vascular endothelial growth factor C. |
Tumor microenvironment and VEGF-C
The comparison OS between low and high numbers of Tr1 and CD4 T cells showed that the levels of Tr1 and CD4 T cells had an observable impact on OS of LUAD patients (Fig. 4a, b). However, no correlation between Tr1 or CD4 cells and VEGF-C was found in Figure 4c, d.
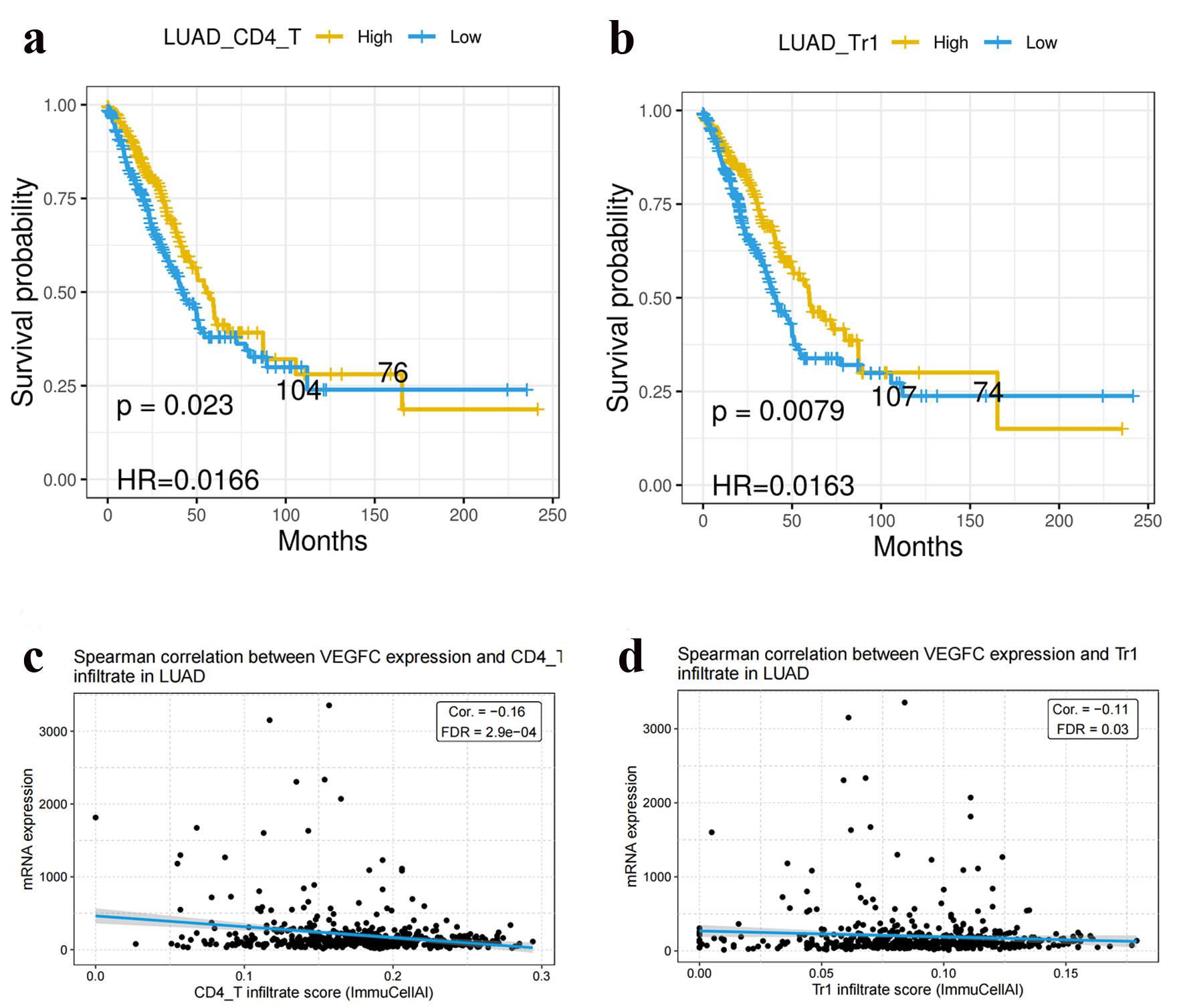 Click for large image | Figure 4. Tr1 cells (a) and CD4 T cells (b) were correlated with OS in LUAD. Tr1 cells (c) and CD4 T cells (d) were not correlated with VEGF-C expression in LUAD. LUAD: lung adenocarcinoma; VEGF-C: vascular endothelial growth factor C. |
VEGF-C expression and predicted drug response for LUAD
To identify effective anti-cancer drugs in LUAD, we used a GSCA server to predict drug responses based on VEGF-C mRNA expression (Fig. 5a, b). Pifithrin-mu showed potential antitumor activity in LUAD (Fig. 5b). As shown in Figure 5a, VEGF-C was positively or negatively correlated with the sensitivity to several chemotherapeutic drugs commonly used in LUAD, like 5-fluorouracil and TGX221. As shown in Figure 5b, VEGF-C was positively correlated with the activity of several chemotherapeutic drugs commonly used in LUAD, such as BI-2536 and BRD-A94377914.
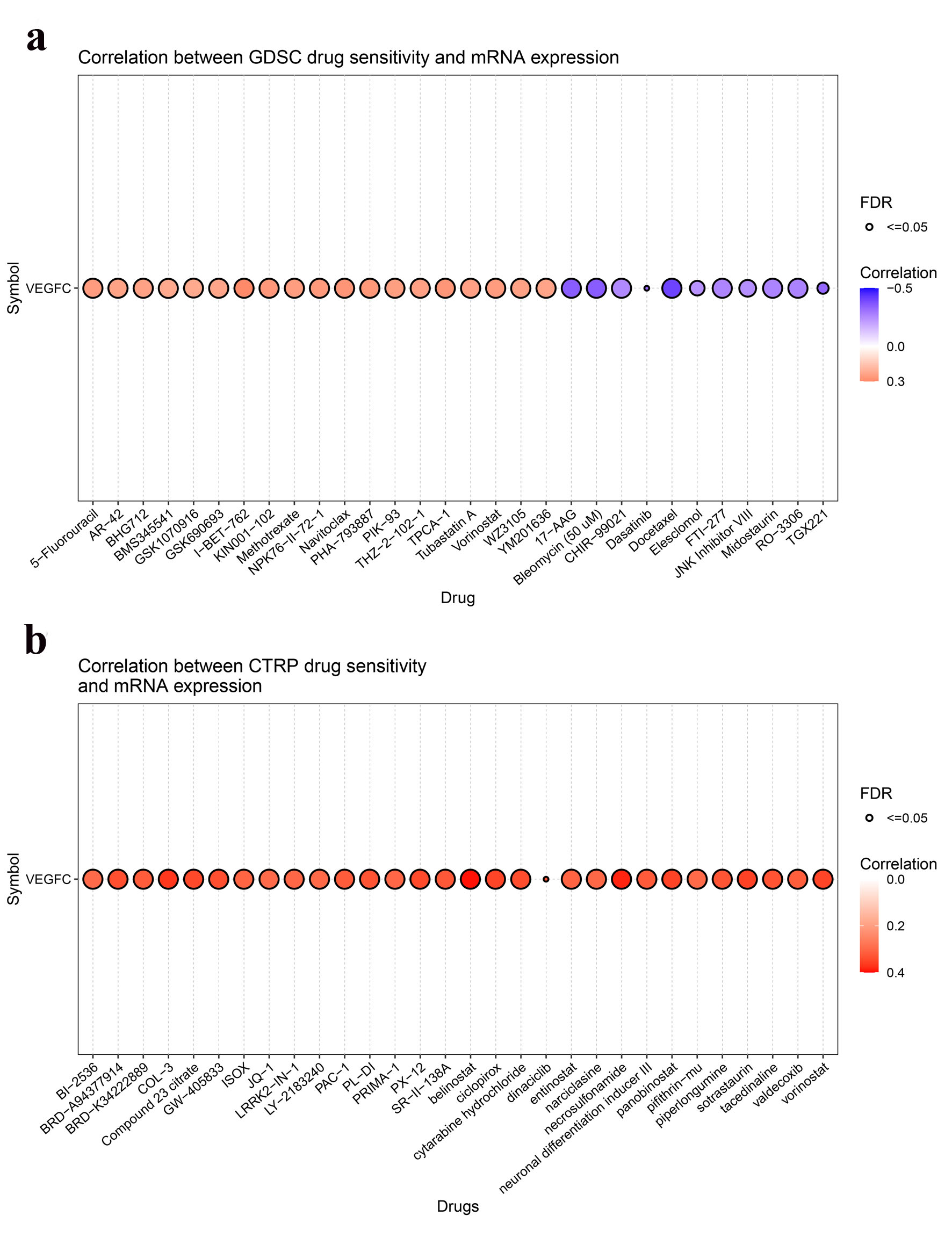 Click for large image | Figure 5. Correlation between VEGF-C expression and drugs sensitivity from GDSC (a) and CTRP (b). VEGF-C: vascular endothelial growth factor C. |
| Discussion | ▴Top |
The application of biomarkers can improve the accuracy of LUAD screening and diagnosis, a vital asset for clinical practice [19]. VEGF-C is particularly important for understanding LUAD because of its angiogenic and lymphangiogenic ability to mediate tumor cell dissemination [20]. GEPIA, UALCAN, TCGAportal, OncoLnc, LCE, GeneMANIA, Metascape, ImmuCellAI, and GSCA online databases were utilized to explore the association of the levels of VEGF-C mRNA expression and outcomes in patients with LUAD. Our study demonstrated that VEGF-C was involved in prognostic related molecular mechanisms in LUAD.
In our analysis, the expression level of VEGF-C mRNA was significantly higher in normal lung tissue than in LUAD tissue and different stages of LUAD patients had significantly different levels of VEGF-C mRNA expression. What’s more, OS of LUAD with low VEGF-C mRNA expression was significantly increased compared to LUAD with high VEGF-C mRNA expression. A meta-analysis conducted among 21 studies also showed that high VEGF-C mRNA expression was associated with inferior LUAD OS (HR = 1.23, 95% CI, 1.15 - 1.32). These results revealed that the expression levels of VEGF-C mRNA might be a valuable prognostic biomarker in LUAD.
VEGF-C can affect the immune system such that tumor cells may escape and stimulate uncontrolled cell proliferation and tumor formation [21]. However, although survival significance of Tr1 cells and CD4 cell infiltrations were observed in this study, our analysis does not find any correlation between CD4+ T cells and Tr1 cells and VEGF-C. Similar findings with our study, Wakabayashi et al have found that CD4+ T cells in cancer stroma were associated with a favorable prognosis in human non-small cell lung cancer (NSCLC) [22]. Similarly, another study has observed that CD4+ T-cell infiltration correlated with a favorable prognosis was observed in sarcoma [23], although this phenomenon was correlated with poor survival in breast cancers [24]. Bonnal et al found that the EOMES+ Tr1-like cells not only correlate with the progression of NSCLC but are also associated with response to programmed death-1 (PD-1) immunotherapy [25].
There was a significant association between VEGF-C expression and mutant TP53 and NF1. Fukuyama et al also observed that TP53 has been shown to be a negative prognostic marker for LUAD [26]. Missense mutant in the TP53 tumor-suppressor gene might drive programmed death ligand-1 (PD-L1) expression through BCL2L1/JAK3/STAT1 pathway [27]. In advanced LUAD patients, TMB was higher in patients with TP53-mutant group than TP53 wild group [27]. TMB is a predictive biomarker in patients with LUAD treated with anti-PD-1/CTLA-4 combination immunotherapy [28]. TP53 was the most frequently mutated gene in NSCLCs harboring NF1 mutation [29]. Inhibition of vascular endothelial growth factor receptor 3 (VEGFR3), a receptor of VEGF-C, could upregulate p53 protein expression in A549 cells. Upregulation of p53 protein expression after VEGFR3 inhibition decreased chemotherapy resistance and improved chemosensitivity [30].
Furthermore, GeneMANIA database analysis showed that VEGF-C interacted with genes including FLT4, VEGFD, KDR, and others. VEGF was the mainly differentially expressed gene by GO analysis, and focal adhesion signaling pathway was mostly involved in the LUAD by KEGG analysis. Function analysis showed that VEGF-C was associated with EGFR TKI resistance. Moehler et al conducted a phase I study to test the efficacy of afatinib, an EGFR TKI, in chemotherapy naive patients with advanced cholangiocarcinoma [31]. Before administration of afatinib, responders had a lower serum level of VEGF-C than non-responders; after therapy, responders demonstrated a decrease of VEGF-C compared to non-responders [31]. Apatinib, a highly selective VEGFR2 inhibitor, significantly potentiated the antitumor effect of gefitinib in NSCLC with T790M-related EGFR TKI resistance [32]. VEGF-C may activate autocrine VEGFR2 signaling to promote cell survival in glioblastoma [33]. What’s more, we observed that the sensitivity of 5-fluorouracil was positively correlated with VEGF-C, and the sensitivity of TGX221 was negatively correlated with VEGF-C. The activity of BI-2536 and BRD-A94377914 was positively correlated with VEGF-C. These findings may need to be validated in further studies with larger sample sizes and prospective study designs.
Our study also has several limitations that need to be considered. First, it was a retrospective study to evaluate the impact of VEGF-C mRNA on outcome of LUAD patients, and further prospective studies are needed to confirm its prognostic role. Second, due to lack of detailed information, some clinicopathological features (such as smoking and drinking status, family history, lymphovascular invasion, tumor size, and so on) may have had an impact on VEGF-C expression. Third, molecular functions and biological effects of VEGF-C in LUAD should be intensively studied in future investigations.
In conclusion, high VEGF-C mRNA expression predicts poor LUAD prognosis. VEGF-C may also be used as a biomarker for immunotherapy. In addition, our study shows that it is possible to identify the optimal LUAD population for anti-VEGF-C therapy.
Acknowledgments
None to declare.
Financial Disclosure
This work was supported in part by a grant from Natural Science Foundation of Jiangsu Science and Technology Department (No. BK20201504) and a grant from Intra-hospital Fund of Jiangsu Provincial Hospital of Traditional Chinese Medicine Development (No. Y20030, Y19064).
Conflict of Interest
The authors declare that they have no competing interests.
Informed Consent
Not applicable.
Author Contributions
QW and JZ designed the study. TP, TL, YW, HH, and XZ collected the data and carried out the analysis. SC, TP and XW wrote the manuscript. All authors read and approved the final manuscript.
Data Availability
The datasets used and/or analyzed during the current study are available from the online databases.
| References | ▴Top |
- Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, Znaor A, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941-1953.
doi pubmed - Kuhn E, Morbini P, Cancellieri A, Damiani S, Cavazza A, Comin CE. Adenocarcinoma classification: patterns and prognosis. Pathologica. 2018;110(1):5-11.
- Ruiz-Patino A, Castro CD, Ricaurte LM, Cardona AF, Rojas L, Zatarain-Barron ZL, Wills B, et al. EGFR Amplification and Sensitizing Mutations Correlate with Survival in Lung Adenocarcinoma Patients Treated with Erlotinib (MutP-CLICaP). Target Oncol. 2018;13(5):621-629.
doi pubmed - Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA. 2019;322(8):764-774.
doi pubmed - Zhan P, Wang J, Lv XJ, Wang Q, Qiu LX, Lin XQ, Yu LK, et al. Prognostic value of vascular endothelial growth factor expression in patients with lung cancer: a systematic review with meta-analysis. J Thorac Oncol. 2009;4(9):1094-1103.
doi pubmed - Jiang H, Shao W, Zhao W. VEGF-C in non-small cell lung cancer: meta-analysis. Clin Chim Acta. 2014;427:94-99.
doi pubmed - Yang Y, Maimaitiyiming X, Jin C, Ahan N, Guo R, Peng C. Influence of Heparanase and VEGF-C mRNA Expressions in Lung Cancer. Indian J Surg. 2015;77(6):477-480.
doi pubmed - Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98-W102.
doi pubmed - Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649-658.
doi pubmed - http://www.tcgaportal.org.
- Anaya J. Guide to using OncoLnc, a new TCGA data portal. Figshare. 2016.
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523.
doi pubmed - Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18-27.
doi pubmed - Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2:e67.
doi - Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214-W220.
doi pubmed - Cai L, Lin S, Girard L, Zhou Y, Yang L, Ci B, Zhou Q, et al. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38(14):2551-2564.
doi pubmed - Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, Guo AY. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv Sci (Weinh). 2020;7(7):1902880.
doi pubmed - Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771-3772.
doi pubmed - Bustos Garcia de Castro A, Ferreiros Dominguez J, Delgado Bolton R, Fernandez Perez C, Cabeza Martinez B, Garcia Garcia-Esquinas M, Carreras Delgado JL. PET-CT in presurgical lymph node staging in non-small cell lung cancer: the importance of false-negative and false-positive findings. Radiologia. 2017;59(2):147-158.
doi pubmed - Das S, Ladell DS, Podgrabinska S, Ponomarev V, Nagi C, Fallon JT, Skobe M. Vascular endothelial growth factor-C induces lymphangitic carcinomatosis, an extremely aggressive form of lung metastases. Cancer Res. 2010;70(5):1814-1824.
doi pubmed - Wang CA, Tsai SJ. The non-canonical role of vascular endothelial growth factor-C axis in cancer progression. Exp Biol Med (Maywood). 2015;240(6):718-724.
doi pubmed - Wakabayashi O, Yamazaki K, Oizumi S, Hommura F, Kinoshita I, Ogura S, Dosaka-Akita H, et al. CD4+ T cells in cancer stroma, not CD8+ T cells in cancer cell nests, are associated with favorable prognosis in human non-small cell lung cancers. Cancer Sci. 2003;94(11):1003-1009.
doi pubmed - Bi Q, Liu Y, Yuan T, Wang H, Li B, Jiang Y, Mo X, et al. Predicted CD4(+) T cell infiltration levels could indicate better overall survival in sarcoma patients. J Int Med Res. 2021;49(1):300060520981539.
doi pubmed - Matkowski R, Gisterek I, Halon A, Lacko A, Szewczyk K, Staszek U, Pudelko M, et al. The prognostic role of tumor-infiltrating CD4 and CD8 T lymphocytes in breast cancer. Anticancer Res. 2009;29(7):2445-2451.
- Bonnal RJP, Rossetti G, Lugli E, De Simone M, Gruarin P, Brummelman J, Drufuca L, et al. Clonally expanded EOMES(+) Tr1-like cells in primary and metastatic tumors are associated with disease progression. Nat Immunol. 2021;22(6):735-745.
doi pubmed - Fukuyama Y, Mitsudomi T, Sugio K, Ishida T, Akazawa K, Sugimachi K. K-ras and p53 mutations are an independent unfavourable prognostic indicator in patients with non-small-cell lung cancer. Br J Cancer. 1997;75(8):1125-1130.
doi pubmed - Sun H, Liu SY, Zhou JY, Xu JT, Zhang HK, Yan HH, Huan JJ, et al. Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. EBioMedicine. 2020;60:102990.
doi pubmed - Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843-852.e844.
doi pubmed - Redig AJ, Capelletti M, Dahlberg SE, Sholl LM, Mach S, Fontes C, Shi Y, et al. Clinical and molecular characteristics of NF1-mutant lung cancer. Clin Cancer Res. 2016;22(13):3148-3156.
doi pubmed - Li Y, Weng Y, Zhong L, Chong H, Chen S, Sun Y, Li W, et al. VEGFR3 inhibition chemosensitizes lung adenocarcinoma A549 cells in the tumor-associated macrophage microenvironment through upregulation of p53 and PTEN. Oncol Rep. 2017;38(5):2761-2773.
doi pubmed - Moehler M, Maderer A, Ehrlich A, Foerster F, Schad A, Nickolay T, Ruckes C, et al. Safety and efficacy of afatinib as add-on to standard therapy of gemcitabine/cisplatin in chemotherapy-naive patients with advanced biliary tract cancer: an open-label, phase I trial with an extensive biomarker program. BMC Cancer. 2019;19(1):55.
doi pubmed - Li F, Zhu T, Cao B, Wang J, Liang L. Apatinib enhances antitumour activity of EGFR-TKIs in non-small cell lung cancer with EGFR-TKI resistance. Eur J Cancer. 2017;84:184-192.
doi pubmed - Michaelsen SR, Staberg M, Pedersen H, Jensen KE, Majewski W, Broholm H, Nedergaard MK, et al. VEGF-C sustains VEGFR2 activation under bevacizumab therapy and promotes glioblastoma maintenance. Neuro Oncol. 2018;20(11):1462-1474.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


