| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 14, Number 1, February 2023, pages 84-93
The Impact of DTYMK as a Prognostic Marker in Colorectal Cancer
Abdulaziz A. Aloliqia, h, Abdul-Fattah Fararjehb, h, i, Ali Al-Khaderc, d, Ezidin Kaddumie, Alaa Abdulaziz Eisaf, Weam Jaradatg
aDepartment of Medical Biotechnology, College of Applied Medical Sciences, Qassim University, Buraydah, Saudi Arabia
bDepartment of Medical Laboratory Sciences, Faculty of Science, Al-Balqa Applied University, Al-salt, Jordan
cDepartment of Pathology and Forensic Medicine, Faculty of Medicine, Al-Balqa Applied University, Al-salt, Jordan
dDepartment of pathology, Al-Hussein Salt Hospital, Al-salt, Jordan
eDepartment of Basic Medical Sciences, Faculty of Medicine, Al-Balqa Applied University, Al-salt, Jordan
fDepartment of Medical Laboratories Technology, College of Applied Medical Sciences, Taibah University, Medina, Saudi Arabia
gDepartment of Medical Laboratory Sciences, Faculty of Graduate Study, Al-Balqa Applied University, Al-Salt, Jordan
hThese authors contributed equally to this article.
iCorresponding Author: Abdul-Fattah Fararjeh, Department of Medical Laboratory Sciences, Faculty of Science, Al-Balqa Applied University, Al-salt, Jordan
Manuscript submitted January 21, 2023, accepted February 8, 2023, published online February 26, 2023
Short title: Prognostic Marker DTYMK in CRC
doi: https://doi.org/10.14740/wjon1571
| Abstract | ▴Top |
Background: Overexpression of deoxythymidylate kinase (DTYMK) has been associated with more aggressiveness and pathological behaviors in hepatocellular carcinoma (HCC) and non-small cell lung cancer (NSCLC). However, the expression of DTYMK and its prognostic significance in colorectal cancer (CRC) patients are yet unknown. The goal of this study was to investigate the DTYMK immunohistochemistry reactivity in CRC tissues and to see how it correlated with various histological and clinical features as well as survival.
Methods: Several bioinformatics databases and two tissue microarrays (TMAs) of 227 cases were used in this study. Immunohistochemistry assay was used to study the protein expression of DTYMK.
Results: Based on the GEPIA, UALCAN, and Oncomine databases, DTYMK expression has increased in tumor tissues at both RNA and protein levels in colorectal adenocarcinoma (COAD) compared to normal tissues. A high DTYMK H-score was found in 122/227 (53%) of the cases, whereas a low DTYMK H-score was found in 105/227. The age at diagnosis (P = 0.036), stage of the disease (P = 0.038), and site of origin (P = 0.032) were all linked to a high DTYMK H-score. Patients with high level of DTYMK had bad overall survival. Interestingly, high DTYMK protein level was associated with PSM2 (P = 0.002) and MSH2 (P = 0.003), but not with MLH2 or MSH6.
Conclusion: This is the first study to cover the expression and prognostic significance of DTYMK in CRC. DTYMK was upregulated in CRC and could be considered as a prognostic biomarker.
Keywords: Colorectal cancer; DTYMK; Prognosis; H-score
| Introduction | ▴Top |
In the United States, colorectal cancer (CRC) is the third most common cancer diagnosed both in women and men, next to prostate cancer (PCa) in men and breast cancer (BC) in women. There are more than 1.9 million new CRC cases every year [1]. Around 40% of all CRC cases are diagnosed in the proximal (cecum) colon, with the other 22% and 28% including the distal colon as well as rectum, respectively [2]. Nevertheless, depending on age and gender, there may be differences in the location of origin. Despite the fact that the incidence and fatality rates of CRC have dropped as a result of improved cancer screening techniques, it is expected that 149,500 new cases will be diagnosed in 2021 in United States, with an estimated 52,980 people dying from the disease according to American Cancer Society [3].
CRC has been linked to a variety of demographic, behavioral, and environmental factors that caused molecular and cellular modification of CRC [4]. The significant risk factors include irritable bowel illness (IBD), CRC history in first-degree relatives, body mass index (BMI), physical activity, cigarette smoking, as well as red meat, fruit, and vegetable consumption [4]. Colonoscopy is still the gold standard for detecting CRC. Computed tomography (CT) scan of the abdomen, chest, and also pelvis with contrast is required before any treatment to stage the patient’s CRC. The TNM classification system is used to stage primary tumor size (T), regional lymph node (N), and distant metastasis (M) [5]. Despite the presence of several tumor markers such as carcinoembryonic antigen which can be raised in CRC, this is not a specific diagnostic biomarker for the disease. Therefore, identifying a novel prognostic and potential diagnostic for CRC patients is urgently needed.
Cell replication requires DNA synthesis, which is especially important in tumor cells. Several cancer types are widely treated with therapeutic drugs that target deoxy-ribonucleotide triphosphate production and metabolism [6]. During a de novo process, deoxythymidine-5′-monophosphate (dTMP) is produced by thymidylate synthase which adds methyl group to deoxyuridine-5′-monophosphate [7]. The salvage process generates dTMP by phosphorylating thymidine with thymidine kinase (TK). Deoxythymidylate kinase (DTYMK) gene is located on chromosome 2 at the 2q37 position and belongs to thymidylate kinase proteins family. The phosphorylation of dTMP to create deoxythymidine diphosphate (dTDP) can be catalyzed by DTYMK. Furthermore, it is the initial step in the formation of deoxythymidine triphosphate (dTTP), which is a crucial component for synthesis of DNA, that combines both salvage and de novo processes [8].
DTYMK is upregulated in a variety of malignancies in humans, including malignant rhabdoid tumors of the kidney [9], acute lymphoblastic leukemia (ALL) [10], and pancreatic cancer [11]. According to Wang et al [12], DTYMK levels were raised in non-small cell lung cancer (NSCLC) using immunohistochemistry (IHC) analysis. Increased expression of DTYMK was found to be strongly linked to NSCLC patients’ aggressive tumor development, including advanced tumor status, increased lymph nodal status, a high TNM stage, and a worse overall survival (OS) [12]. More recently, DTYMK has been shown to be a poor prognostic biomarker in hepatocellular carcinoma (HCC) [13]. Another study demonstrated the prognostic significance of 70 genes in BC. DTYMK was found to be one of the most upregulated gene signatures associated with poor OS in BC [14].
There have been no investigations performed on DTYMK in CRC regarding the expression or the prognostic significance yet. In this study, we investigated the expression of DTYMK at mRNA and protein levels using several bioinformatics databases and IHC assay.
| Materials and Methods | ▴Top |
Reagents and tissue microarrays (TMAs)
Two TMA sections of 227 cases were purchased from US Biomax (CO1021a and CO1251). Antibody against DTYMK was purchased from Abcam (anti-DTYMK antibody, cat. number: ab154867). Rabbit HRP/DAB IHC detection kit was purchased from Abcam (cat. number: ab64261).
IHC staining and evaluation
To determine the clinical importance of DTYMK expression in CRC, IHC staining for DTYMK was done on TMA CRC tissues of 227 cases. The TMA slides were prepared for antigen retrieval by heating them to 93 °C for 30 min using a citric acid antigen retrieval solution (pH 6.0). After that, the sections were incubated for 10 min with 3% H2O2. The two slides were treated in a humidified atmosphere at 4 °C for 24 h with antibody produced against DTYMK (1:200 dilution). The IHC detection kit was then used to do the IHC staining. DTYMK immunostaining was assessed by specialized pathologists. The percentage and intensity of positively stained tumor cells were used to assess staining. The cells were classified as DTYMK-positive if the cytoplasm of the tumor cells was slightly stained with DTYMK antibody. The following equation was used to get the H-score: H-score = Pi (I + 1), where I is the stained tumor cell intensity (0, 1+, 2+ and 3+), and Pi is the percentage of the tumor cells that stained for each intensity [15]. The H-score mean was used as the cutoff value, which was set at 6 out of 12. DTYMK expression was considered high in cases of an H-score greater than 6.
Expression profile of DTYMK in CRC
The DTYMK gene expression pattern at RNA level for the pairs of tumor vs. normal tissues in colorectal and other types of malignancies was studied using bioinformatics methods. GEPIA is a website that provides RNA data from the GTEx and TCGA investigations, which has over 9,700 and 8,487 for tumors and normal samples, respectively. Customizable features offered by GEPIA include tumor/normal differential expression analysis and cancer type-specific profiling. The RNA-Seq datasets GEPIA used is based on the UCSC Xena project in order to locate the DTYMK gene, and we used the expression DIY functionality. After that, we chose profile option from the expression DIY menu and added CRC (COAD) to the datasets list. Then, automatically, matched normal data, log scale, and Jitter size were all established [16].
GEO dataset website [17] was used for searching studies on COAD. A validated GEO microarray dataset (GSE156720) including three pairs of colorectal tumor and normal tissues was obtained from GEO dataset [18].
UALCAN is a web-based resource for studying cancer OMICS data that is comprehensive, user-friendly, and interactive. UALCAN is a detailed, viewer, and an interactive website resource for evaluating cancer OMICS data. It allows users to search for biomarkers, to perform in silico validation of interested genes, and to create graphs and also plots depicting gene level profiles [19].
Over 700 datasets in various types of cancers are available in the Oncomine database [20], all of which are of high quality and have a substantial study cohort. Oncomine was used to study the expression of DTYMK at RNA level in CRC cohort studies.
Using data from the TCGA-COAD cohort, the Cancer Genomics Browser [21] at UCSC (University of California in Santa Cruz, CA, USA) was utilized to evaluate the association between DTYMK at the mRNA level with CRC and understand the DTYMK gene’s prognostic value in COAD.
Clinical characterization of patients with CRC
To further investigate the relationship of DTYMK protein levels with clinical and pathological features in CRC, two TMAs (n = 227) were chosen. A total of 105 (46%) of the 227 cases were under the age of 59, while 122 (54%) were beyond the age of 60 (Table 1). More than half of the cases were males (139/227 (60%)), while females accounted for 91/227 (40%). According to the TNM staging system, tumor size (T) was divided into four categories: for T1, the tumor is only in the inner layer (n = 1); for T2, the tumor grows into muscle (n = 37); for T3, the tumor grows into the outer lining but not grown through it (n = 119); and for T4, the tumor spread (n = 65 cases). The following criteria were used to establish the lymph node status: N0 means no lymph node metastasis (n = 155), N1 means metastasis in 1 - 3 regional lymph nodes (n = 47) and N2 means metastasis in 4 - 9 regional lymph nodes (n = 20). In none of the cases, there were any distant metastases. Furthermore, the clinical stage was classified as follows: stage I (n = 36), stage II (n = 1,419), stage III (n = 64), and stage IV (n = 3). Tumor grade included low grade (n = 44), moderate grade (n = 139), and high grade (n = 33).
 Click to view | Table 1. Clinical and Pathological Features for the 227 Colorectal Cancer (COAD) Patients |
Statistical analysis
SPSS version 19 was used to evaluate the TMAs and COAD data. Student’s t-test and Pearson Chi-squared test were used to analyze the expression level of DTYMK at RNA and protein level in normal vs. tumor COAD cases, in addition to association of the DTYMK with COAD clinical and pathological parameters. To compare survival curves plotted using Kaplan-Meier analysis, the log-rank test was utilized. P < 0.05 and P < 0.001 were considered as statistically significant.
| Results | ▴Top |
Expression level of DTYMK gene in COAD
We compared the levels of DTYMK expression in normal and malignant tissues in COAD using GEPIA based on profile gene analysis. DTYMK gene was overexpressed in tumor tissues more than normal tissues in breast cancer (BRCA) (tumor (T) = 1,085/normal (N) = 112), COAD (T = 275/N = 41), liver hepatocellular carcinoma (LIHC) (T = 369/N = 50)), lung squamous cell carcinoma (LUSC) (T = 483/N = 59), and prostate adenocarcinoma (PRAD) (T = 495/N = 52). Notably, the expression of DTYMK was higher in COAD in comparison to the other types of cancers, which has an average around 45 transcripts per million TPM (Fig. 1a). We next obtained an RNA seq file based on transcriptome analysis to verify our findings. DTYMK levels in tumors were substantially greater than in normal tissues (P = 0.009) (Fig. 1b). In addition, as shown, we used the UALCAN website to assess the mRNA and protein levels of DTYMK in large study samples (Fig. 1c, d) with P-value of 1.6 ×10-12 and 9.0 × 10-39, respectively. Primary tumor tissues had higher quantities of mRNA and protein than normal tissues. We also looked at the degree of DTYMK expression in these datasets using different COAD cohort studies from the Oncomine database: rectal adenocarcinoma vs. normal, Gaedcke colorectal/colorectal carcinoma vs. normal, Graudens/colorectal carcinoma vs. normal, Hong colorectal/colorectal adenocarcinoma vs. normal, Skrzypczak colorectal/colorectal carcinoma vs. normal, Skrzypczak colorecta. DTYMK RNA expression was high in majority of the study cohorts, as seen in Figure 1e, f. Overall, tumor tissue showed a higher level of DTYMK than normal tissue, according to these data.
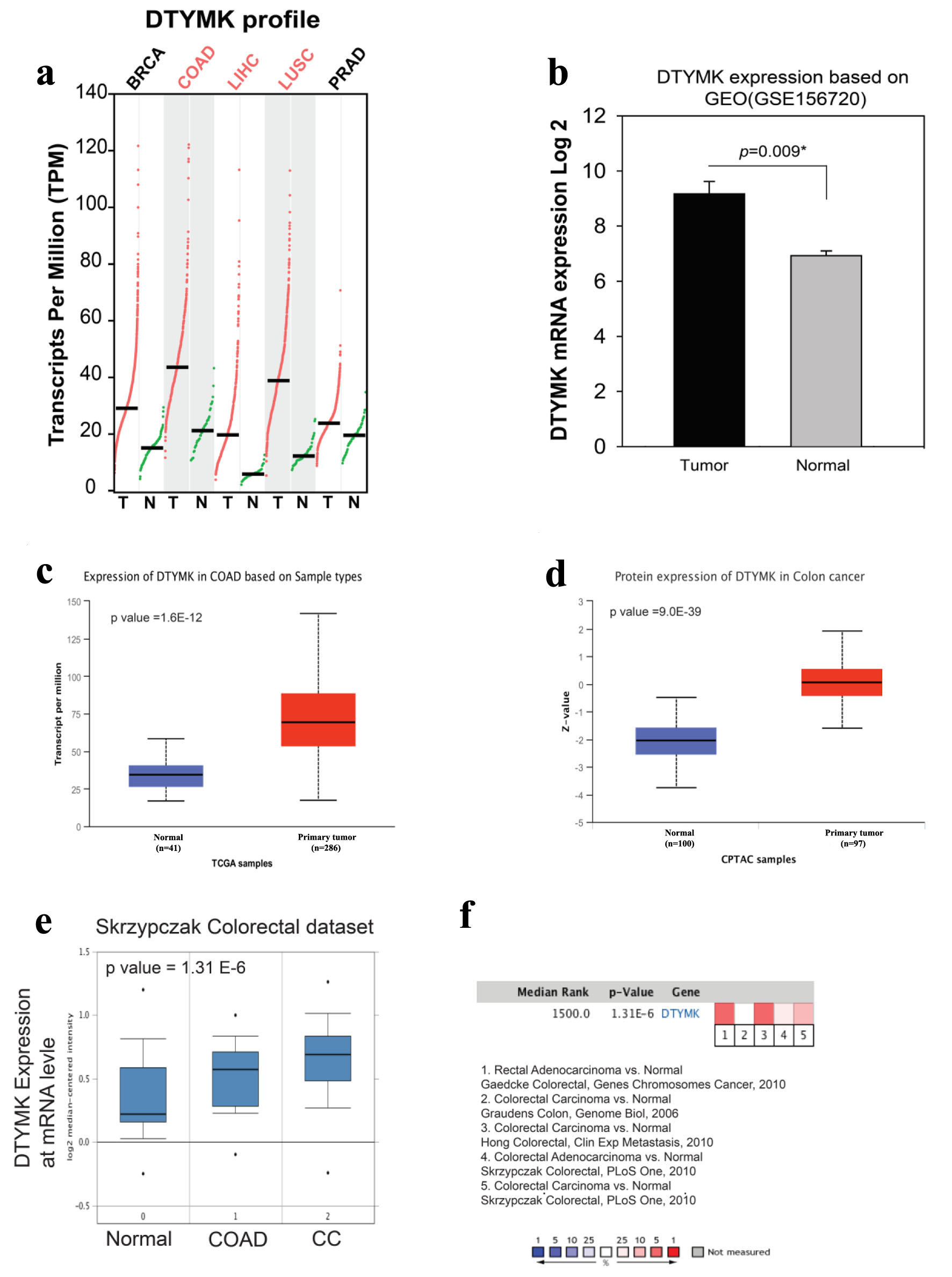 Click for large image | Figure 1. High expression level of DTYMK in colorectal cancer (COAD). (a) DTYMK gene expression was upregulated in several types of human malignancies especially in COAD. (b) Using GEO dataset, analysis of DTYMK expression in three pairs of tumors vs. normal of CRC patients. (c, d) Using UALCAN website, analysis of DTYMK at mRNA and protein levels. (e, f) Using Oncomine database, analysis of DTYMK in several COAD study cohorts. *P value < 0.05 was considered to be significant [16]. CRC: colorectal cancer; DTYMK: deoxythymidylate kinase; COAD: colorectal adenocarcinoma. |
DTYMK positivity and clinical and pathological parameters
The COAD cases were evaluated according to expression of DTYMK (negative 0, weak 1+, moderate 2+ and strong 3+) (Fig. 2a). The percentage of positive and the intensity of DTYMK staining were used to determine the H-score (Table 2). Patients were categorized into two groups depending on their H-score, with the mean H-score being 9.6 out of 12. A representative image in Figure 2b shows the high H-score versus low H-score cases, which indicated the cytoplasmic expression for DTYMK.
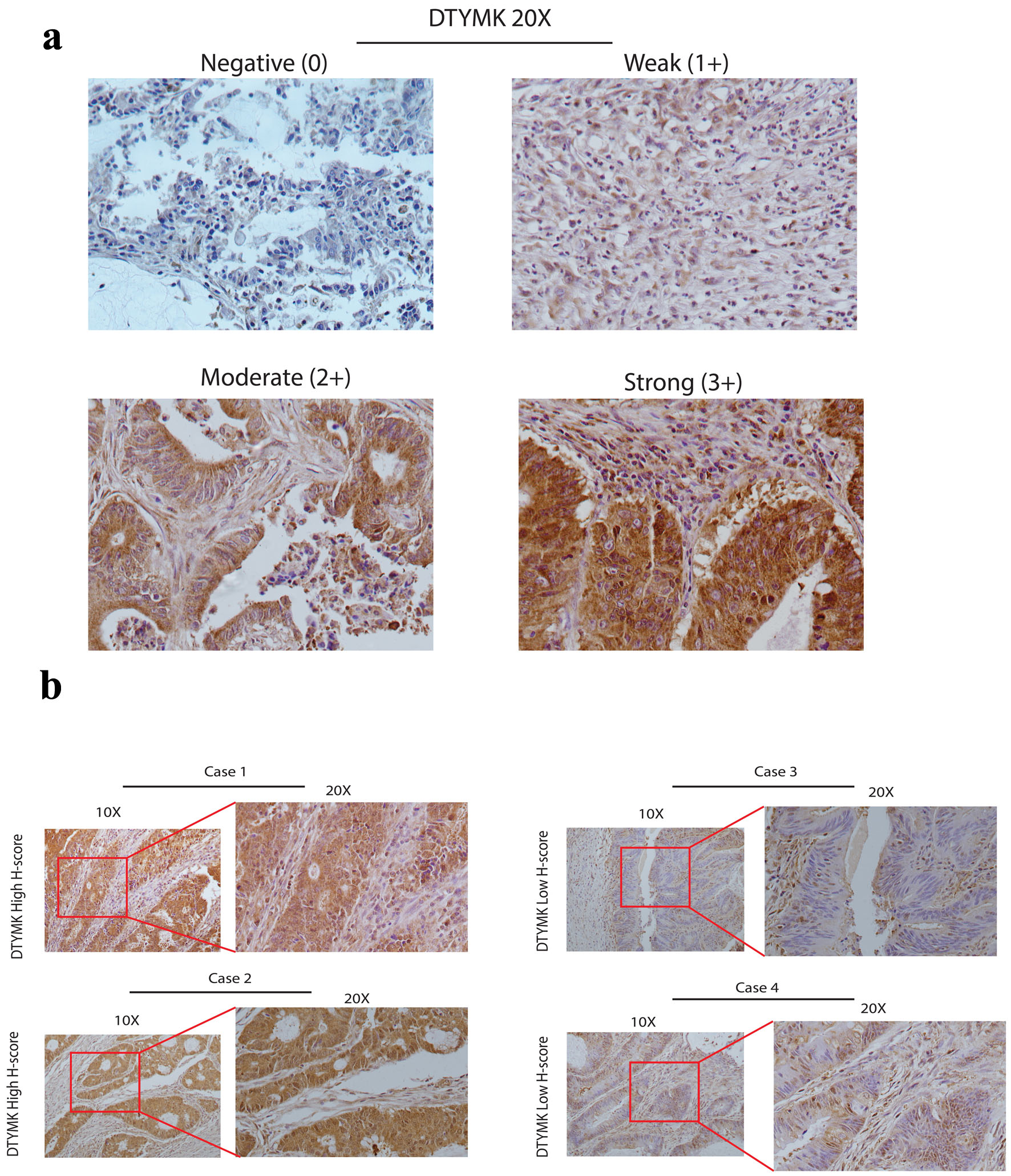 Click for large image | Figure 2. DTYMK protein expression by IHC. (a) These images show the DTYMK IHC grading system (original magnification × 20). Negative (0), weak (1+), moderate (2+), and strong (3+) grading systems were constructed based on DTYMK staining intensity. (b) In four cases, illustrative photographs depict examples of high DTYMK H-score and low DTYMK low H-score (original magnification was × 10 and × 20). DTYMK: deoxythymidylate kinase; IHC: immunohistochemistry. |
 Click to view | Table 2. The Findings of the Scores Obtained for DTYMK Protein Expression |
The details of DTYMK H-score with clinical and pathological parameters are shown in Table 3. DTYMK was linked to the patient’s age, tumor stage, and location of origin. There was no link between DTYMK and the patient’s gender, tumor grade, tumor size, or lymph node status.
 Click to view | Table 3. Comparison of Patients With Low Versus High DTYMK H-Score in Their Colorectal Adenocarcinomas Pathological Variables (n = 227) |
High DTYMK H-score was more associated with patient’s age more than 60 years old (122/227) (P = 0.036). Moreover, DTYMK was associated with earlier stage of the tumor (P = 0.038) and the primary origin site, left and right colon has the highest expression levels (P = 0.032).
The correlation between DTYMK protein expression and the mismatch repair (MMR) genes
CRC is caused by germline mutations in the DNA MMR genes MSH2, MSH6, or MLH1. These mutations have an autosomal dominant inheritance pattern [22, 23]. PMS2 encodes a protein that is required for MMR; nevertheless, mutations in this gene have only been recorded in exceedingly rare situations [24-26]. In this study, we examined the correlation between the MMR proteins IHC results with DTYMK expression (Fig. 3a-d). We found that high DTYMK H-score is strongly associated with high level of PSM2 (P = 0.003) and MSH2 (P = 0.002), but not with MSH6 and MLH1. These results may indicate that co-expression level of DTYMK with PMS2 and MSH2 could work together in CRC patients.
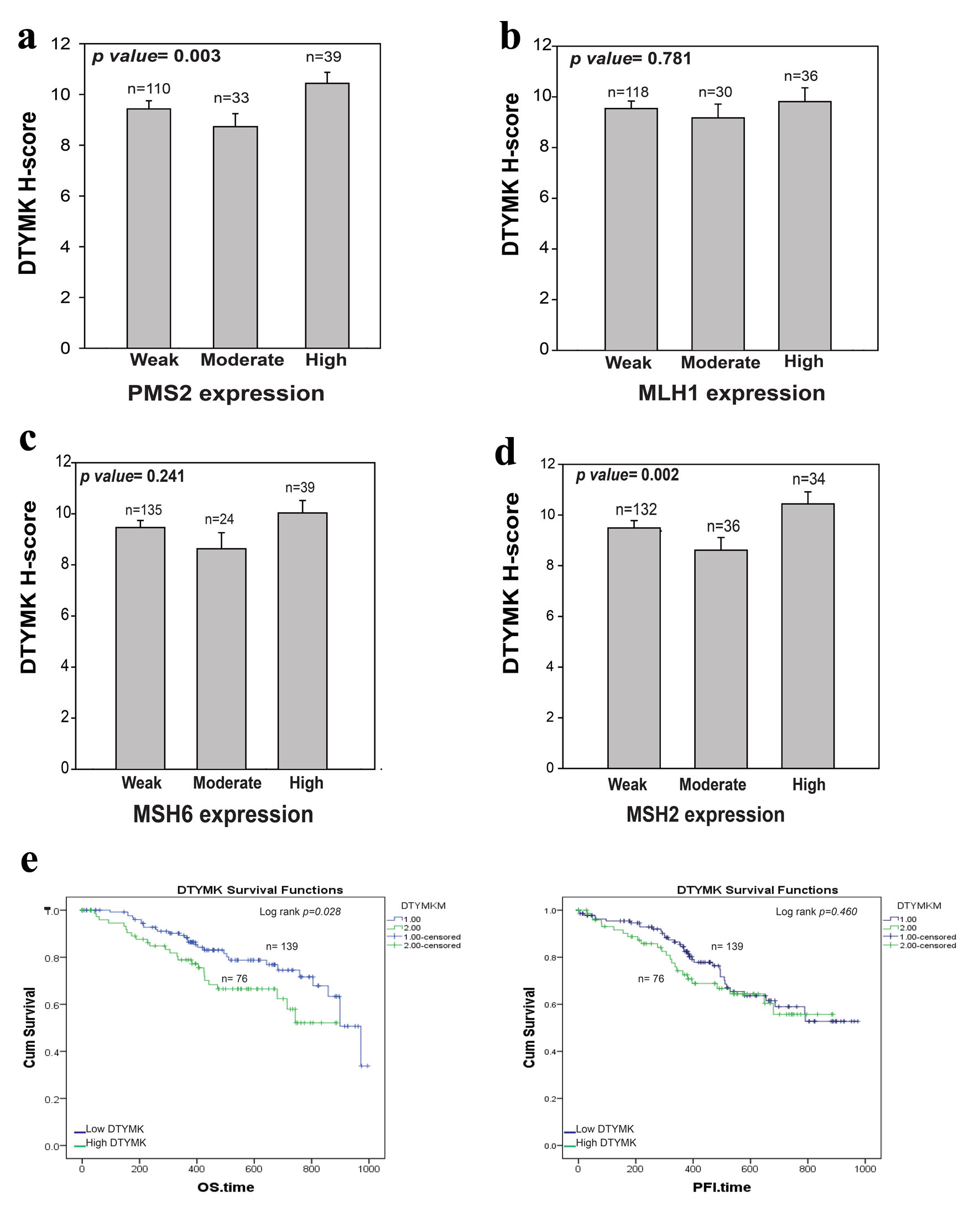 Click for large image | Figure 3. Correlation between DTYMK and MMR proteins. (a-d) Analysis of the association between DTYMK IHC and PMS2, MLH1, MSH6 and MSH2 IHC results, respectively. P value < 0.05 was considered to be significant. (e) The overall survival (OS) and progression-free interval (PFI) analysis for high DTYMK vs. low DTYMK mRNA levels. Log rank P value < 0.05 was considered to be significant. DTYMK: deoxythymidylate kinase; IHC: immunohistochemistry; MMR: mismatch repair. |
The prognostic significance of DTYMK in COAD
TCGA-COAD data were downloaded from Xenobrowser website. The survival outcomes were analyzed based on DTYMK mRNA level. DTYMK gene expression was separated into two groups based on the mean level of DTYMK gene levels, the mean expression was 9.86 (Log2 of DTYMK mRNA level). Patients above the mean had high DTYMK expression. We found that patients that had high level of DTYMK (n = 76) have poor overall survival (OS) in comparison to those with low level of DTYMK (n = 139) (P = 0.028) (Fig. 3e). Patients with high level of DTYMK has bad progression free survival (PFI) for less than 500 days of the disease. However, no differences were found between low vs. high after 600 days of the disease.
| Discussion | ▴Top |
The main purpose of this study was to assess DTYMK IHC reactivity in CRC (COAD) patients retrospectively and to investigate the link between the degree of its expression and histological and clinical factors that may influence patients’ prognosis. For measuring DTYMK expression, we utilized a DTYMK IHC H-score (percentage multiplied by intensity) that has been previously used in various BC and CRC studies [15, 27, 28].
In our study, we revealed that DTYMK RNA level in COAD tumor was considerably elevated than in surrounding normal tissues by evaluating data using UALCAN, GEPIA, GEO and Oncomine databases. In this study, we showed that DTYMK could be used as a prognostic biomarker in COAD patients. The TCGA data were used to validate DTYMK’s prognostic value, and Kaplan-Meier analysis was used to estimate survival outcomes. Early-stage COAD patients and those over 60 years old were more likely to have elevated DTYMK expression. Moreover, left and right hemicolons have shown to be at high DTYMK protein level. DTYMK was found to be increased in NSCLC and HCC, which is consistent with our findings [12, 13, 29, 30]. To validate these results, 227 CRC cases were selected for IHC study. Immunochemical results showed that more than half of the patients (122/227, 53%) had a high H-score.
The role of DTYMK in cancers has been investigated previously in HCC. It was identified that DTYMK acts as an oncogene, and knocking down DTYMK in HCC cells reduced cell growth and disturbed the cell cycle. The expression of cell cycle proteins which are implicated in the G0/G1 and S phases was drastically reduced after inhibition of DTYMK in HCC cells. Furthermore, after loss of DTYMK function, Hep3B and Huh7 HCC cells lines developed to be more susceptible to sorafenib and oxaliplatin [29].
In CRC, one of the standard medications used in chemotherapy regimens is 5-fluorouracil (5-FU). In two different 5-FU resistance CRC cell lines, DTYMK was found to be upregulated in both cell lines, which may be linked to medication resistance in CRC cells [31]. Furthermore, it was demonstrated that, by using two HCT116 colon cancer cell lines with p53+, + and another p53-, -, loss of DTYMK expression using lentivirus short hairpin RNA (shRNA) led to decrease in the dTTP pool without affecting p53 expression levels [32]. In addition, it has shown that inhibited DTYMK p53-, - colon cells enhanced doxorubicin sensitivity. After exposure to low-dose doxorubicin, the DNA damage response was augmented and apoptotic induction was accelerated, resulting in cell death [32]. There have been no reports that state the level of DTYMK expression or the association between DTYMK level and the clinic-pathological aspects of patients.
Apoptosis evasion, which is a hallmark of cancer, is critical for cancer survival, cell growth, therapy-resistance, in addition to metastasis in CRC [33]. Microsatellite instability (MSI) has been revealed in CRC to be immunogenic with substantial lymphocytic infiltrate due to enhanced mutational signatures. MSI CRC is defined by the absence or high level of one or more MMR proteins which include MSH2, MLH1, PMS2 and MSH6 [34]. IHC identification of DNA MMR proteins in CRC screening for CRC is being studied extensively. In our study, and in consistency with another study [35], 48-59% of the cases showed low expression of MMR proteins and almost 17% of the cases showed high expression of MMR proteins. Interestingly, high protein expression of DTYMK was associated with PSM2 and MSH2 but not with MSH6 or MLH1. DTYMK based on our preliminary study (not shown) has a strong protein-protein interaction with RAD51 which has a vital role in DNA repair by homologous recombination [36]. No direct interaction has been found between DTYMK and MMR proteins except through RAD51. A previous study demonstrated that RAD51 was overexpressed in CRC and associated with tumor stage for patients with CRC as we found for DTYMK [37]. Further studies are needed to clarify the correlation between MMR protein and DTYMK and RAD51 in patients with CRC.
More recently, stomatin-like 2 (STOML2) was shown to be highly overexpressed in CRC, and its presence was linked to a poor prognosis [38]. Based on the yeast 2 hybrid and computational biology analysis, the authors screened the most interacted proteins with STOML2. DTYMK was one of the most similar proteins in terms of signaling pathways and biological reactions, which may indicate an important role of DTYMK in CRC [38].
There are a few things to mention. Firstly, the importance of DTYMK expression levels as a biomarker needs to be established by a prospective validation study because this research was retrospective. Second, based on our findings, DTYMK was linked to poor OS in CRC, which we believed to be age-related. Additionally, the link between early stage and DTYMK expression does not support the result. Therefore, more investigations are required.
Conclusion
We detect in this study high level of DTYMK at mRNA and protein levels in CRC patients more than normal tissues. High DTYMK H-score was detected in 53% of CRC and low H-score in 47%. It has been discovered that patients with a high DTYMK level had a worse OS rate in comparison to those with a low level. DTYMK may be a novel, potent prognostic biomarker, as well as a therapeutic target in CRC, according to the study, which recommends for more researches.
Acknowledgments
None to declare.
Financial Disclosure
The researchers would like to thank the Deanship of Scientific Research, Qassim University, for funding the publication of this project.
Conflict of Interest
The authors have nothing to disclose related to this work.
Informed Consent
Not applicable because of the database study.
Author Contributions
AAA, AFF, and AAK were in charge of scoring, statistical analysis, and interpretation of the data. The project was designed by AAA, AFF, and EK, who also wrote the manuscript. The manuscript text was written with the help of AAE and WJ. The manuscript was revised by all writers.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
| References | ▴Top |
- Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet. 2005;365(9454):153-165.
doi pubmed - Cheng L, Eng C, Nieman LZ, Kapadia AS, Du XL. Trends in colorectal cancer incidence by anatomic site and disease stage in the United States from 1976 to 2005. Am J Clin Oncol. 2011;34(6):573-580.
doi pubmed - Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7-33.
doi pubmed - Johnson CM, Wei C, Ensor JE, Smolenski DJ, Amos CI, Levin B, Berry DA. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control. 2013;24(6):1207-1222.
doi pubmed - Buccafusca G, Proserpio I, Tralongo AC, Rametta Giuliano S, Tralongo P. Early colorectal cancer: diagnosis, treatment and survivorship care. Crit Rev Oncol Hematol. 2019;136:20-30.
doi pubmed - Kohnken R, Kodigepalli KM, Wu L. Regulation of deoxynucleotide metabolism in cancer: novel mechanisms and therapeutic implications. Mol Cancer. 2015;14:176.
doi pubmed - Van Triest B, Pinedo HM, Giaccone G, Peters GJ. Downstream molecular determinants of response to 5-fluorouracil and antifolate thymidylate synthase inhibitors. Ann Oncol. 2000;11(4):385-391.
doi pubmed - Hebrard C, Cros-Perrial E, Clausen AR, Dumontet C, Piskur J, Jordheim LP. Bacterial deoxyribonucleoside kinases are poor suicide genes in mammalian cells. Nucleosides Nucleotides Nucleic Acids. 2009;28(11):1068-1075.
doi pubmed - Nagata T, Takahashi Y, Ishii Y, Asai S, Sugahara-Kobayashi M, Nishida Y, Murata A, et al. Molecular genetic alterations and gene expression profile of a malignant rhabdoid tumor of the kidney. Cancer Genet Cytogenet. 2005;163(2):130-137.
doi pubmed - Meyer LH, Eckhoff SM, Queudeville M, Kraus JM, Giordan M, Stursberg J, Zangrando A, et al. Early relapse in ALL is identified by time to leukemia in NOD/SCID mice and is characterized by a gene signature involving survival pathways. Cancer Cell. 2011;19(2):206-217.
doi pubmed - Nagayoshi Y, Nakamura M, Matsuoka K, Ohtsuka T, Mori Y, Kono H, Aso T, et al. Profiling of autoantibodies in sera of pancreatic cancer patients. Ann Surg Oncol. 2014;21(Suppl 3):S459-465.
doi pubmed - Wang W, Guo ZH, Lu XP, Liao DJ, Peng GL, Xu X, He JX. Elevated expression of DTYMK is associated with poor prognosis in patients with Non-small cell lung cancer. International Journal of Clinical & Experimental Medicine. 2016;9(11):22027-22033.
- Guo Y, Luo W, Huang S, Zhao W, Chen H, Ma Y, Ye M, et al. DTYMK expression predicts prognosis and chemotherapeutic response and correlates with immune infiltration in hepatocellular carcinoma. J Hepatocell Carcinoma. 2021;8:871-885.
doi pubmed - Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, Robertson JF, et al. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2007;26(10):1507-1516.
doi pubmed - Fararjeh AS, Chen LC, Ho YS, Cheng TC, Liu YR, Chang HL, Chang HW, et al. Proteasome 26S subunit, non-ATPase 3 (PSMD3) regulates breast cancer by stabilizing HER2 from degradation. Cancers (Basel). 2019;11(4):527.
doi pubmed - Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98-W102.
doi pubmed - https://www.ncbi.nlm.nih.gov/gds.
- https://www.ncbi.nlm.nih.gov/geo/.
- http://ualcan.path.uab.edu/.
- http://www.oncomine.org.
- https://xenabrowser.net/.
- Hung S, Saiakhova A, Faber ZJ, Bartels CF, Neu D, Bayles I, Ojo E, et al. Mismatch repair-signature mutations activate gene enhancers across human colorectal cancer epigenomes. Elife. 2019;8:40760.
doi pubmed - Wang C, Wang Y, Hughes KS, Parmigiani G, Braun D. Penetrance of colorectal cancer among mismatch repair gene mutation carriers: a meta-analysis. JNCI Cancer Spectr. 2020;4(5):pkaa027.
doi pubmed - Chou A, Fraser T, Ahadi M, Fuchs T, Sioson L, Clarkson A, Sheen A, et al. NTRK gene rearrangements are highly enriched in MLH1/PMS2 deficient, BRAF wild-type colorectal carcinomas-a study of 4569 cases. Mod Pathol. 2020;33(5):924-932.
doi pubmed - Yilmaz A, Mirili C, Bilici M, Tekin SB. Colorectal cancer in Lynch syndrome associated with PMS2 and MSH6 mutations. Int J Colorectal Dis. 2020;35(2):351-353.
doi pubmed - Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, Bannwart F, et al. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128(5):1160-1171.
doi pubmed - Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, Attar A, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16(1):102-108.
doi pubmed - Fararjeh AS, Tu SH, Chen LC, Liu YR, Lin YK, Chang HL, Chang HW, et al. The impact of the effectiveness of GATA3 as a prognostic factor in breast cancer. Hum Pathol. 2018;80:219-230.
doi pubmed - Sun F, Liu Y, Gong T, Pan Q, Xiang T, Zhao J, Tang Y, et al. Inhibition of DTYMK significantly restrains the growth of HCC and increases sensitivity to oxaliplatin. Cell Death Dis. 2021;12(12):1093.
doi pubmed - Zhou T, Qin R, Shi S, Zhang H, Niu C, Ju G, Miao S. DTYMK promote hepatocellular carcinoma proliferation by regulating cell cycle. Cell Cycle. 2021;20(17):1681-1691.
doi pubmed - de Angelis PM, Fjell B, Kravik KL, Haug T, Tunheim SH, Reichelt W, Beigi M, et al. Molecular characterizations of derivatives of HCT116 colorectal cancer cells that are resistant to the chemotherapeutic agent 5-fluorouracil. Int J Oncol. 2004;24(5):1279-1288.
doi pubmed - Hu CM, Chang ZF. Synthetic lethality by lentiviral short hairpin RNA silencing of thymidylate kinase and doxorubicin in colon cancer cells regardless of the p53 status. Cancer Res. 2008;68(8):2831-2840.
doi pubmed - Liu L, Cao C, Zhu Y, Li D, Feng H, Luo J, Tang Z, et al. Preoperative chemoradiotherapy with 5-fluorouracil and oxaliplatin for locally advanced rectal cancer: long-term results of a phase II trial. Med Oncol. 2015;32(3):70.
doi pubmed - Boland PM, Ma WW. Immunotherapy for colorectal cancer. Cancers (Basel). 2017;9(5):50.
doi pubmed - Shia J, Tang LH, Vakiani E, Guillem JG, Stadler ZK, Soslow RA, Katabi N, et al. Immunohistochemistry as first-line screening for detecting colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: a 2-antibody panel may be as predictive as a 4-antibody panel. Am J Surg Pathol. 2009;33(11):1639-1645.
doi pubmed - Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391(6665):401-404.
doi pubmed - Lee JH, Bae AN, Jung AS. Clinicopathological and Prognostic Characteristics of RAD51 in Colorectal Cancer. Medicina (Kaunas). 2020;56(2):48.
doi pubmed - Ma W, Chen Y, Xiong W, Li W, Xu Z, Wang Y, Wei Z, et al. STOML2 interacts with PHB through activating MAPK signaling pathway to promote colorectal Cancer proliferation. J Exp Clin Cancer Res. 2021;40(1):359.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


