| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 14, Number 3, June 2023, pages 205-223
Comprehensive Analysis Reveals the Potential Roles of Transcription Factor Dp-1 in Lung Adenocarcinoma
aSchool of Law and Criminal Justice, East China University of Political Science and Law, Songjiang University Town, Shanghai 201620, China
bShanghai Institute of Blood Transfusion, Shanghai Blood Center, Shanghai 200051, China
cCorresponding Author: Rongna Ma, Shanghai Institute of Blood Transfusion, Shanghai Blood Center, Shanghai 200051, China
Manuscript submitted April 3, 2023, accepted May 22, 2023, published online June 11, 2023
Short title: Potential Roles of TFDP1 in LUAD
doi: https://doi.org/10.14740/wjon1595
| Abstract | ▴Top |
Background: Transcription factor Dp-1 (TFDP1) was overexpressed and interacted with other genes to impact multiple signaling pathways in various human cancers. However, there is less research about the TFDP1 specific roles in lung adenocarcinoma (LUAD).
Methods: We first explored TFDP1 expression levels and relative diseases from a pan-cancer perspective using the ONCOMINE, TIMER, and Open Targets Platform databases. Then, we used UALCAN, GEPIA 2, TCGA-LUAD data, and Kaplan-Meier plotter to examine TFDP1 clinicopathological features and prognosis in LUAD patients. Genomic alterations and DNA methylation analysis were performed by cBioPortal and MethSurv, respectively. Then, we used a cancer single-cell state atlas (CancerSEA) to find TFDP1 functions at a single-cell resolution. LinkedOmics was used to find TFDP1 coexpressed genes, biological processes, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Then, Gene Set Cancer Analysis (GSCA) was used to examine the drug resistence of TFDP1 in LUAD.
Results: We found that TFDP1 was overexpressed in most human cancers and related to various diseases, including LUAD. Moreover, LUAD patients with high TFDP1 expression levels might be significantly associated with individual cancer stages and have a poor prognosis. Multivariate analysis revealed that the American Joint Committee on Cancer (AJCC) pathologic stage, AJCC stage T, and AJCC stage N were the independent prognostic factors. LUAD patients with TFDP1 alterations suggested poor overall survival (OS), and disease-free survival (DFS), while hypermethylation might lead to a good prognosis. TFDP1 and its coexpressed genes were enriched in multiple signaling pathways and biological processes involved in the cell cycle, spliceosome, and DNA replication. Furthermore, TFDP1 was strongly positively related to the half-maximal inhibitory concentration (IC50) values of multiple drugs.
Conclusions: In summary, TFDP1 was a possible biomarker and potential therapeutic target for LUAD patients.
Keywords: TFDP1; Omics analysis; Prognostic biomarker; Therapeutic target; Lung adenocarcinoma
| Introduction | ▴Top |
Lung cancer is one of the most worldwide malignancies, with 11.4% of 19.3 million new cancer patients in the world in 2020, leading to 1.8 million deaths worldwide and ranking as the primary cause of cancer mortality [1]. Lung cancer causes about 30% of cancer deaths and is China’s most common malignant tumor [2], where lung adenocarcinoma (LUAD) is the most prevalent subtype [3, 4]. Although different therapies have been used in LUAD patients, the survival rate still remains at a low level, which can seriously reduce human life expectancy [5, 6]. Therefore, identifying novel biomarkers for improving the prognosis of LUAD patients is hugely imperative and meaningful.
Transcription factor Dp-1 (TFDP1), also known as DRTF1, DP1, or Dp-1, plays a vital role in various human cancers [7-9]. Melchor et al showed that 13q34 amplification significantly overexpressed TFDP1 protein, and TFDP1 was one of the most potential target genes in human breast cancer [10]. Pellicelli et al revealed that TFDP1 participated in the transcriptional regulation of PITX1 in osteoarthritis [11]. Recent research showed that TFDP1 was identified as a critical colorectal cancer (CRC) molecule and could be directly bound to miR-4711-5p, which provoked G1 arrest in the CRC [12, 13]. TFDP1 was identified as a probable target of 13q34 amplification in hepatocellular carcinomas [14]. Drucker et al showed that karyopherin-α2 imported E2F1/TFDP1 to regulate the protein stathmin in liver cancer [15]. COMMD9 acted on TFDP1 to enhance the p53 signaling pathway in non-small cell lung cancer (NSCLC) [16]. Five genes, including TFDP1, were used to establish a prognostic signature nomogram in LUAD [17]. However, there are few studies on the specific roles and molecular mechanisms of TFDP1 in LUAD.
Therefore, using various databases, we first analyzed TFDP1 expression levels in different human cancers and found TFDP1-related diseases. Then, we explored TFDP1 overexpression associated with clinicopathologic features and prognosis of LUAD patients. We evaluated TFDP1 genomic alterations and DNA methylation in LUAD patients. LinkedOmics was used to explore TFDP1 relative genes and Gene Set Cancer Analysis (GSCA) in LUAD. We further investigated TFDP1 functions at single-cell resolution and the drug sensitivity of TFDP1-coexpressed genes. We systematically analyzed the roles of TFDP1 in LUAD and found that TFDP1 can be a possible prognostic biomarker for treating LUAD patients.
| Materials and Methods | ▴Top |
Expression level analysis
ONCOMINE [18] is an online cancer database, which contains 715 data sets and 86,733 samples [19]. In this study, ONCOMINE was utilized for comparing differences in the TFDP1 transcriptional expression in various human cancers between tumors and normal tissues. These were the thresholds: gene rank: 10%, data type: mRNA, P value: 0.01, and fold change: 1.5.
TIMER [20] is a useful web resource to investigate immune infiltrates across human cancers systematically. Users can input specific parameters to intuitively study the human cancer’s genomic features, clinical, and immunological by dynamically displaying figures [21]. The DiffExp module was employed to explore the differential levels of any gene of interest between cancer tissues and normal tissues accross all The Cancer Genome Atlas (TCGA) tumors, in which statistical significance is evaluated using the Wilcoxon test. We used the DiffExp module of TIMER to verify TFDP1 expression in various human tumors. The expression levels were displayed with (log_2 (TPM)).
UALCAN [22] is an intuitive web tool for studying genes of interest in cancer data (Children Brain Tumor Tissue Consortium (CBTTC), Clinical Proteomics Tumor Analysis Consortium (CPTAC), and TCGA) [23]. Thus, we employed UALCAN to investigate the TFDP1 protein expression differences in the CPTAC database.
Relative diseases analysis
With scoring and ranking target-disease relationships, the Open Targets Platform [24] utilizes drugs, animal models, scientific literatures, genomics, transcriptomics, and genetics to identify drug targets [25]. We used the Open Targets Platform to find TFDP1-associated diseases.
Pathological characteristics analysis
We used UALCAN to analyze the correlation between TFDP1 mRNA expression and individual cancer stages and histological subtypes of LUAD patients. Gene Expression Profiling Interactive Analysis (GEPIA) 2 [26] is a user-friendly web tool for investigating the RNA-Seq levels of 9,736 cancers and 8,587 normal samples in the Genotype-Tissue Expression (GTEx) and the TCGA datasets [27]. We utilized pathological stage plot to profile TFDP1 expression in LUAD stages with a box plot. Then, we utilized SPSS 19.0 (SPSS Inc., Chicago, IL, USA) to perform univariate and multivariate Cox regression analysis for assessing the prognostic indicators of overall survival (OS) in LUAD (using the TCGA-LUAD dataset). With the average TFDP1 mRNA levels, LUAD patients are divided into the high-expression and low-expression groups.
Survival prognosis analysis
Kaplan-Meier plotter [28] is mainly used to evaluating the impact of 54,000 genes on survival values in 21 cancer types (European Genome-phenome Archive (EGA), Gene Expression Omnibus (GEO), and TCGA) [29]. We used the Kaplan-Meier plotter to investigate the association of TFDP1 expression levels with the prognosis values in LUAD patients (OS, first progression survival (FPS), and post progression survival (PPS)).
Genomic alteration analysis
cBioPortal [30] can provide researchers with a high-quality and intuitive online resource to explore multi-dimensional cancer datasets. This study utilized cBioPortal to study TFDP1 genomic alterations in LUAD patients from the TCGA database [31, 32]. GeneMANIA [33] uses 2,830 association network data to study functions of input genes and research other genes related to the input gene sets [34]. We created gene-gene interaction networks for TFDP1, GRK1, LAMP1, RASA3, and ATP4B using GeneMANIA and discovered their associated genes in LUAD patients.
Methylation analysis
MethSurv [35] provides a rapid and intuitive web to perform survival analysis of DNA methylation data in 25 human cancers and 7,358 patients in the TCGA database [36]. MethSurv were employed to study TFDP1 DNA methylation CpG sites in LUAD patients and the association of TFDP1 DNA methylation levels with the LUAD prognosis values.
Single-cell analysis
CancerSEA [37] can comprehensively decode 14 functions from 41,900 single cells spanning 25 different cancers at the single-cell resolution [38]. We used CancerSEA to find which functional states of TFDP1 were related to LUAD.
LinkedOmics analysis
LinkedOmics [39] is an interactive web resource for investigating cancer multi-omics data in 32 types of TCGA cancer and 10 CPTAC cancer cohorts [40]. Firstly, we utilized the LinkFinder module to explore coexpressed genes of TFDP1 in the TCGA LUAD data (Pearson Correlation test). Then, the LinkInterpreter module performed enrichment analysis about differential genes from the LinkFinder results. Gene Set Enrichment Analysis (GSEA) was employed to do gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The rank criterion was as follows: minimum number of genes: 3, false discovery rate (FDR) < 0.01, and simulations: 500.
Drug sensitivity analysis
GSCA [41] was employed to examine the association between TFDP1 expression and drug sensitivity in the Cancer Therapeutics Response Portal (CTRP) and Genomics of Drug Sensitivity in Cancer (GDSC) databases [42]. The scores of the TFDP1-drug pairs were calculated by -log10FDR * |cor|, with the absolute correlation coefficient |cor| > 0.1 and FDR < 0.05. The top 30 ranked drugs were demonstrated by the lollipop plot.
The Institutional Review Board approval and the declaration of ethical compliance with human/animal study are not applicable for this study.
| Results | ▴Top |
TFDP1 expression levels and its relative diseases
To explore differences in TFDP1 transcriptional levels between cancer tissues and normal tissues across various human cancers, we employed ONCOMINE to analyze TFDP1 levels in 20 types of cancer. Compared with normal tissues, TFDP1 mRNA levels were significantly overexpressed in 15 of all 20 cancer types, including lung cancer (Fig. 1a). Then, we studied TFDP1 expression patterns in human cancers using the TIMER database, which contains the RNA-seq data of various human cancers in the TCGA database. TFDP1 expression levels were markedly downregulated in kidney renal clear cell carcinoma (KIRC), thyroid carcinoma (THCA), kidney renal papillary cell carcinoma (KIRP), and kidney chromophobe (KICH), while TFDP1 levels were markedly upregulated in 13 cancers, including LUAD (Fig. 1b).
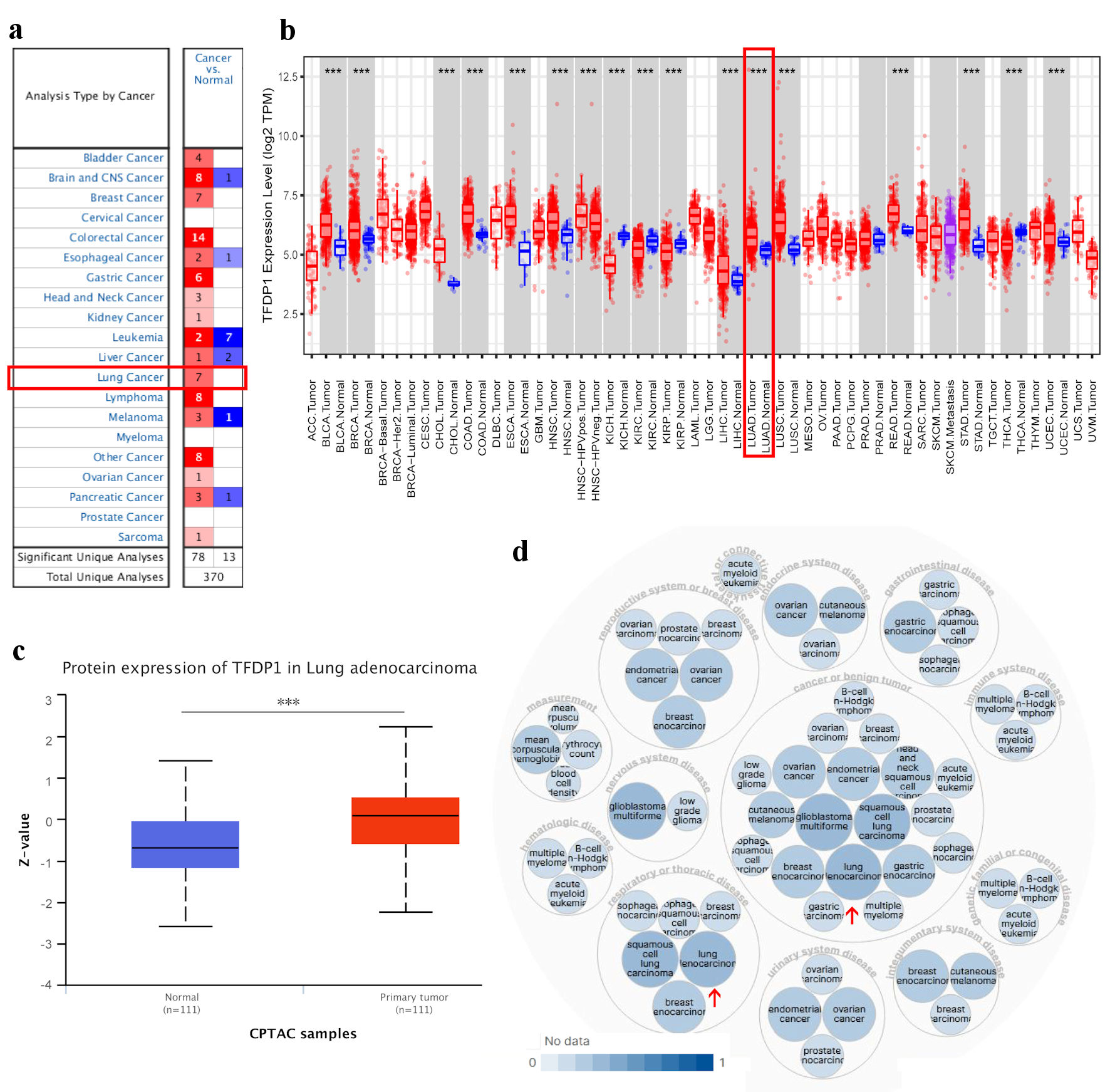 Click for large image | Figure 1. TFDP1 expression in various types of human cancer and its relative diseases. (a) The difference of TFDP1 mRNA levels in cancer tissues and normal tissues (ONCOMINE). (b) The TFDP1 expression difference from the TIMER database. (c) The TFDP1 protein expression differences in the CPTAC database (UALCAN). (d) TFDP1-associated diseases obtained by Open Targets Platform. (***P < 0.001). TFDP1: transcription factor Dp-1; CPTAC: Clinical Proteomics Tumor Analysis Consortium. |
Then, we tried to analyze TFDP1 protein expression levels using the CPTAC database with UALCAN. Compared with normal tissues, the protein expression levels of TFDP1 were significantly upregulated in LUAD tissues (Fig. 1c). In addition, using the Open Targets Platform, we also found that TFDP1 was associated with respiratory (e.g., LUAD), thoracic, reproductive system, breast, gastrointestinal, immune system, and endocrine system diseases (Fig. 1d). These findings revealed that the TFDP1 transcriptional levels were markedly overexpressed in LUAD.
TFDP1 pathological characteristics analysis in LUAD patients
Using the UALCAN and GEPIA 2 databases, we further analyzed the relationship between TFDP1 mRNA expression and the pathological stages in LUAD. We found that TFDP1 mRNA levels were highly related with the cancer stages in LUAD (Fig. 2a). The TFDP1 levels increased with each cancer stage of LUAD, reaching its greatest expression in the stage 4. Similarly, we also found a remarkable correlation (P = 0.00459) of TFDP1 expression with pathological stages using GEPIA 2 (Fig. 2b), in which patients with more advanced pathological stages were inclined to have higher TFDP1 expression. Moreover, TFDP1 was overexpressed in all LUAD histological subtypes compared with normal tissues, and the highest expression levels were found in the lung clear cell adenocarcinoma subtypes. TFDP1 expression was significantly upregulated in the LUAD-not otherwise specified (NOS), LUAD-mixed, lung bronchioloalveolar carcinoma non mucinous (LBC-nonmucinous), lung solid pattern predominant adenocarcinoma, lung acinar adenocarcinoma, and lung papillary adenocarcinoma subtypes (Fig. 2c). Then, Cox univariate analysis revealed that TFDP1 mRNA expression, American Joint Committee on Cancer (AJCC) pathologic stage, AJCC stage T, and AJCC stage N were significantly associated with OS in LUAD (Table 1). Multivariate analysis further confirmed that AJCC pathologic stage, AJCC stage T, and AJCC stage N were the independent prognostic factors in LUAD. These findings revealed a substantial correlation between the TFDP1 mRNA levels and the LUAD pathological phases.
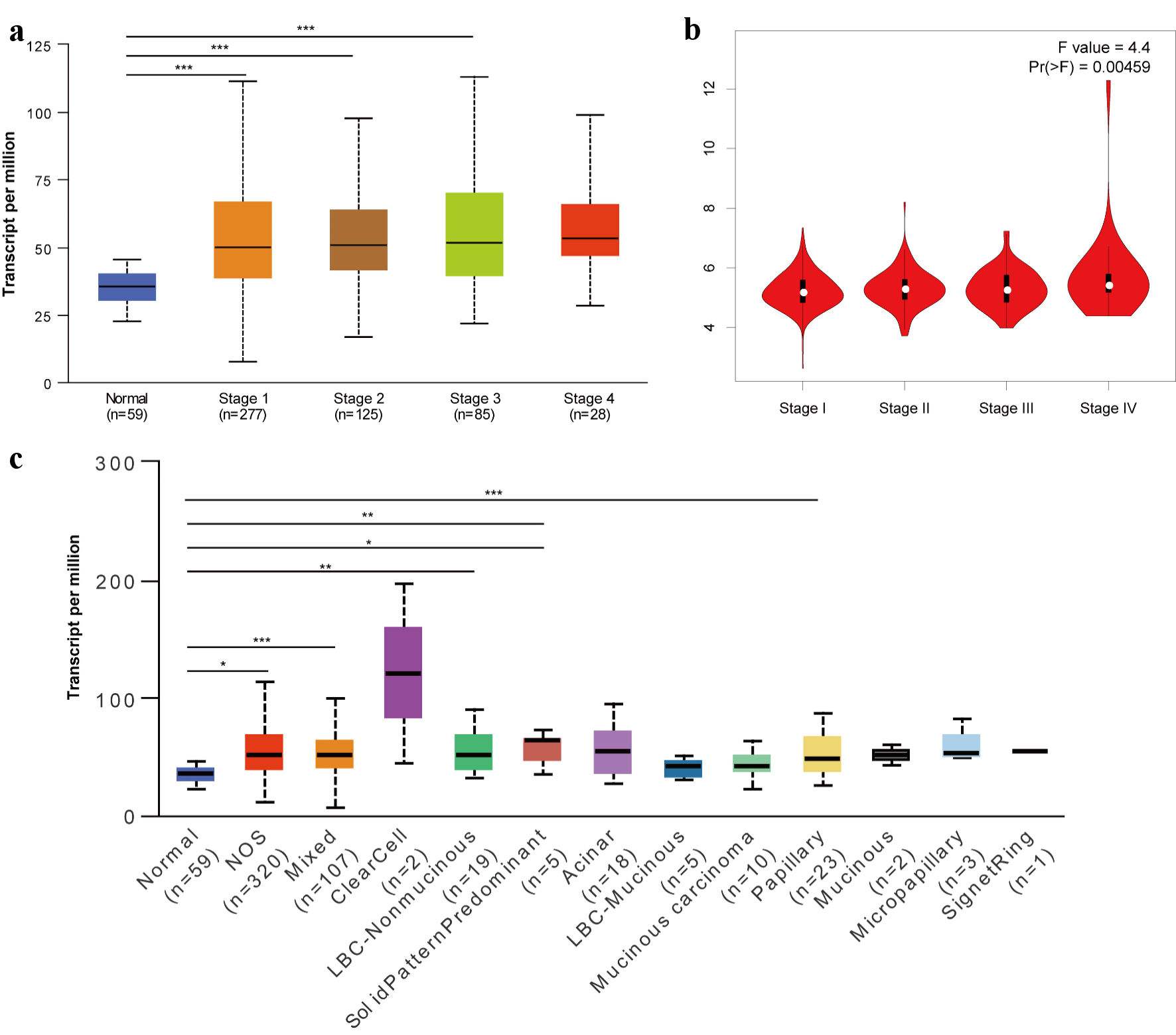 Click for large image | Figure 2. Relationship of TFDP1 with the pathological stages of LUAD patients analyzed by UALCAN (a) and GEPIA 2 (b). (c) TFDP1 expression differences in the LUAD histological subtypes. (*P < 0.05, **P < 0.01, ***P < 0.001). TFDP1: transcription factor Dp-1; LUAD: lung adenocarcinoma. |
 Click to view | Table 1. Univariate and multivariate Cox regression analysis in LUAD |
High TFDP1 levels impacts on the LUAD prognosis
Next, the Kaplan-Meier plotter was employed to investigate whether TFDP1 mRNA levels were associated with LUAD prognosis. As shown in Figure 3, LUAD patients having high TFDP1 expression demonstrated a poor OS (hazard ratio (HR) = 2.43, 95% confidence interval (CI): 1.88 - 3.14), P = 1.8 × 10-12), FPS (HR = 1.72, 95% CI: 1.24 - 2.39, P = 0.0011), and PPS (HR = 1.78, 95% CI: 1.07 - 2.96, P = 0.024). These results indicated that higher TFDP1 levels were associated with worse prognosis and TFDP1 could be a prognostic biomarker in LUAD. These above findings revealed that TFDP1 was crucial to the tumorigenesis and progression of LUAD.
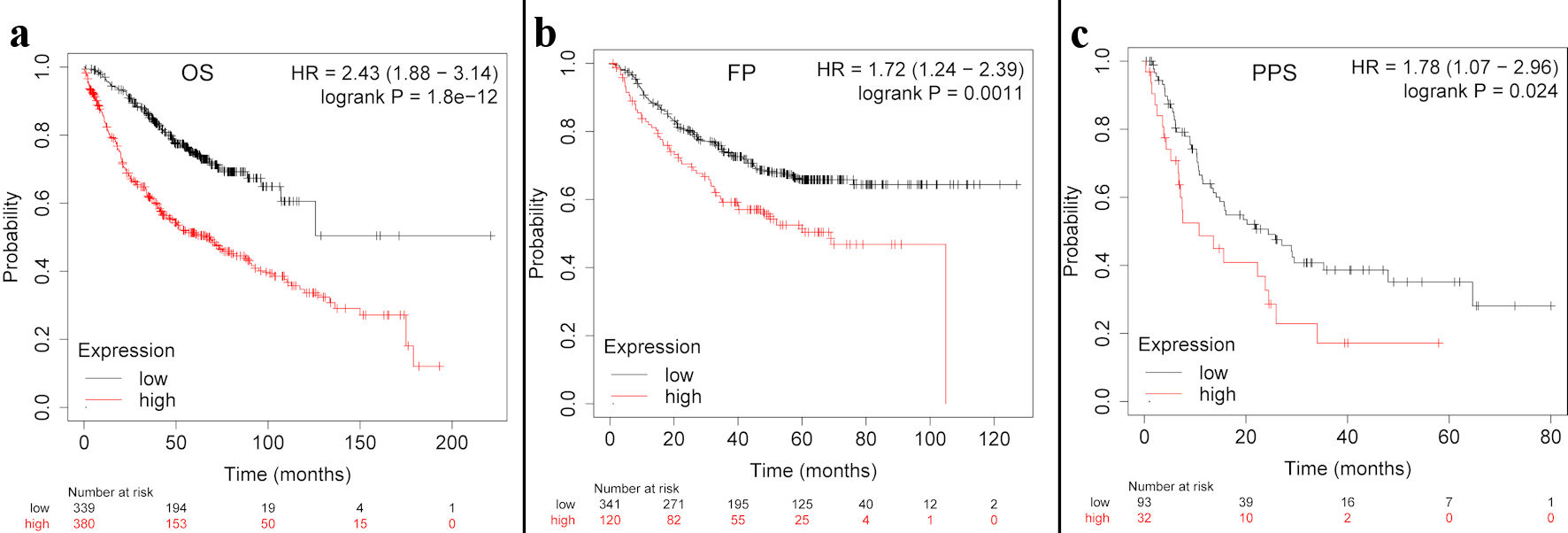 Click for large image | Figure 3. The TFDP1 prognosis in LUAD patients using the Kaplan-Meier plotter. (a)The OS curve of TFDP1 in LUAD. (b) The first progression (FP) curve of TFDP1 in LUAD. (c) The PPS curve of TFDP1 in LUAD. TFDP1: transcription factor Dp-1; LUAD: lung adenocarcinoma; OS: overall survival; PPS: post progression survival. |
Genomic alteration analysis of TFDP1 in LUAD patients
With the TCGA data, we applied cBioPortal to analyze genomic alterations of TFDP1 in LUAD. Figure 4a displayed five types of TFDP1 alterations in various human cancers. Moreover, TFDP1 was altered in 3.89% of 566 LUAD patients, including deep deletion: 1.77% (10 cases), mutation: 1.59% (nine cases), amplification: 0.35% (two cases) and multiple alterations: 0.18% (one case).
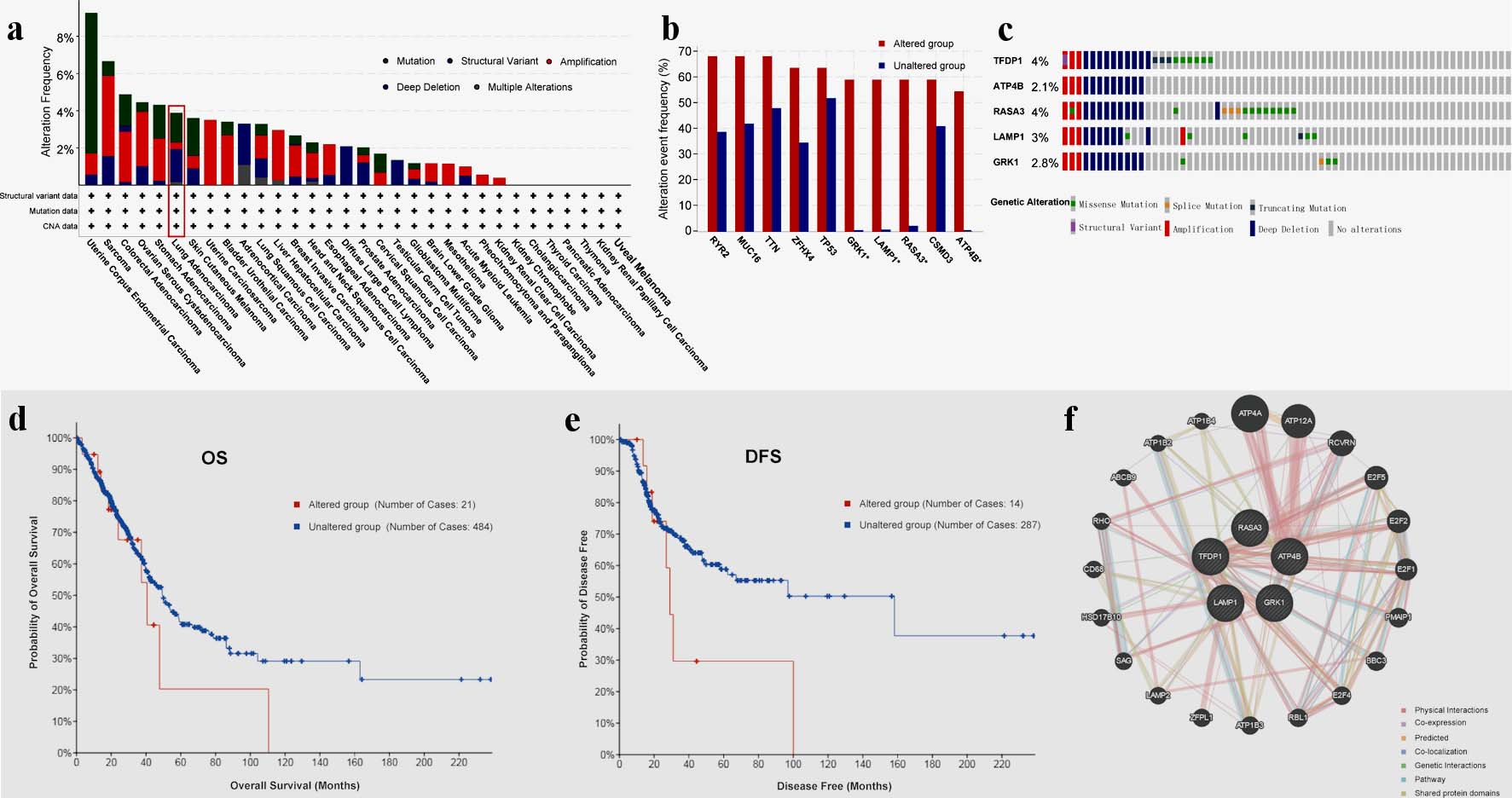 Click for large image | Figure 4. TFDP1 alterations in LUAD patients analyzed by cBioPortal and GeneMANIA. (a) Alteration frequencies of TFDP1 across various human cancers. (b) Alteration frequencies of RYR2, MUC16, TTN, ZFHX4, TP53, GRK1, LAMP1, RASA3, CSMD3, and ATP4B cooccurred with TFDP1 alterations in LUAD. (c) Summary of genetic alterations of TFDP1, ATP4B, RASA3, LAMP1, and GRK1 in LUAD. (d) The OS and (e) the DFS between TFDP1-altered and TFDP1-unaltered groups in LUAD patients. (f) Gene-gene interaction network of TFDP1 and its cooccurred genes. TFDP1: transcription factor Dp-1; LUAD: lung adenocarcinoma; OS: overall survival;DFS: disease-free survival. |
We demonstrated that the 10 genes with the highest alteration frequencies in the LUAD altered groups were RYR2, MUC16, TTN, ZFHX4, TP53, GRK1, LAMP1, RASA3, CSMD3, and ATP4B, cooccurred with TFDP1 modifications (Fig. 4b). Additionally, we found that TFDP1 alterations cooccurred with GRK1, LAMP1, RASA3, and ATP4B (log ratio > 4). Also, GeneMANIA was employed to create a gene-gene interaction network of the five genes (Fig. 4c). Compared with LUAD patients without TFDP1 alteration, TFDP1-altered patients might have a poor OS (Fig. 4d) and disease-free survival (DFS) values (Fig. 4e).
DNA methylation analysis of TFDP1 in LUAD patients
We used MethSurv to analyze TFDP1 DNA methylation in 461 LUAD patients from the TCGA data. The results revealed that TFDP1 at most CpG sites had higher DNA methylation levels (Fig. 5). Moreover, we found that four CpG sites (cg10439691, cg11976193, cg26355737, and cg01595804) were significantly associated with the prognostic values (Table 2). High DNA methylation of CpG cg10439691 and cg11976193 were strongly related to poor prognosis, while hypermethylation of CpG cg26355737 and cg01595804 was strongly related to a good prognostic value in LUAD.
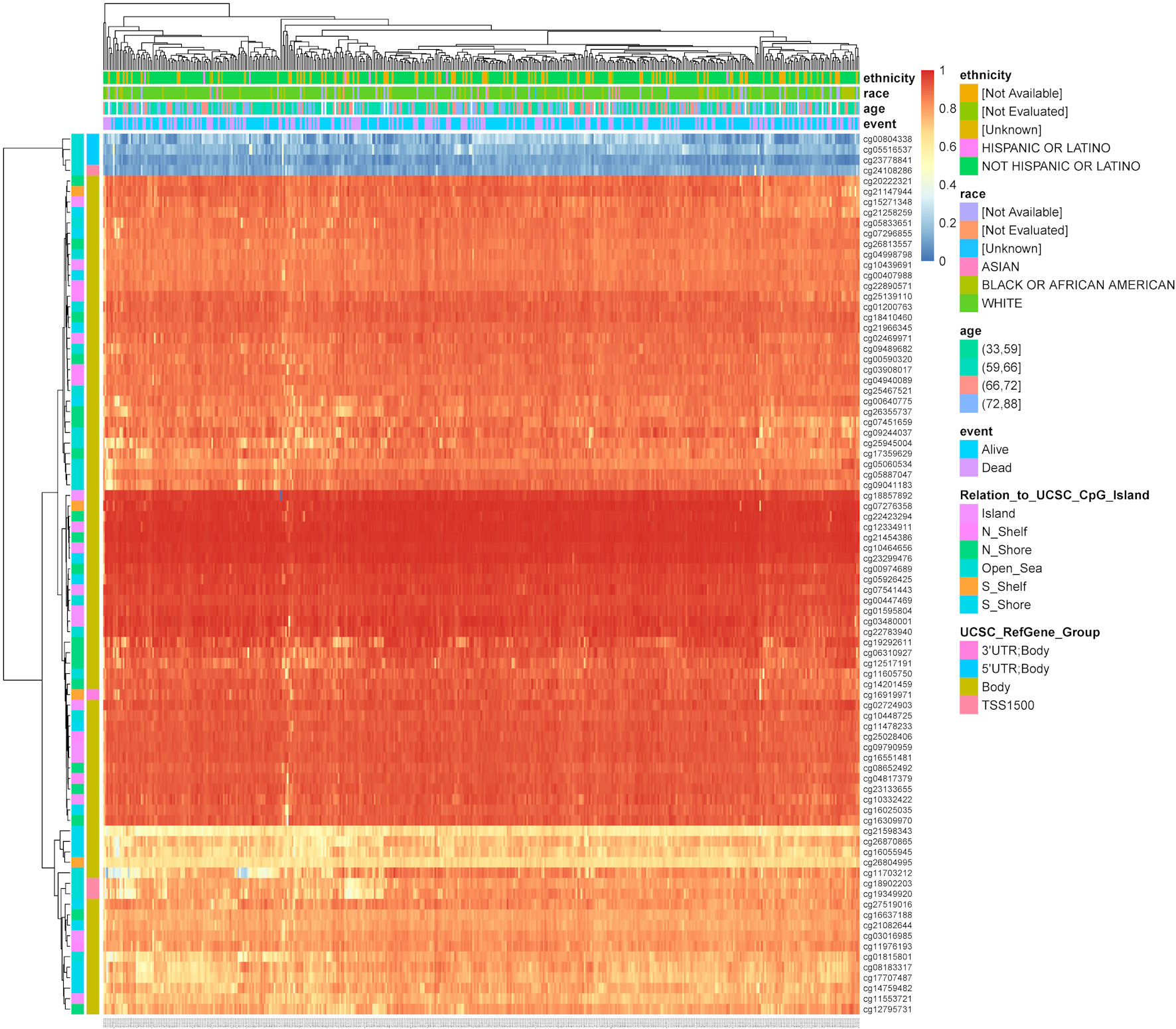 Click for large image | Figure 5. The heatmap shows TFDP1 methylation levels at different CpG sites in LUAD patients (MethSurv). TFDP1: transcription factor Dp-1; LUAD: lung adenocarcinoma. |
 Click to view | Table 2. The Significant Correlation of TFDP1 Methylation at Different CpG Sites With the Prognosis in LUAD |
Functions of TFDP1 in LUAD
To further study TFDP1 functions in LUAD, CancerSEA was employed to investigate the functional heterogeneity from a single-cell resolution. Figure 6a illustrated the correlations between TFDP1 and 14 functional states in LUAD. We found that TFDP1 was positively related to cell cycle (P ≤ 0.01), DNA damage (P ≤ 0.01), proliferation (P ≤ 0.05), and DNA repair (P ≤ 0.05) in the EXP0067 dataset (Fig. 6b-f).
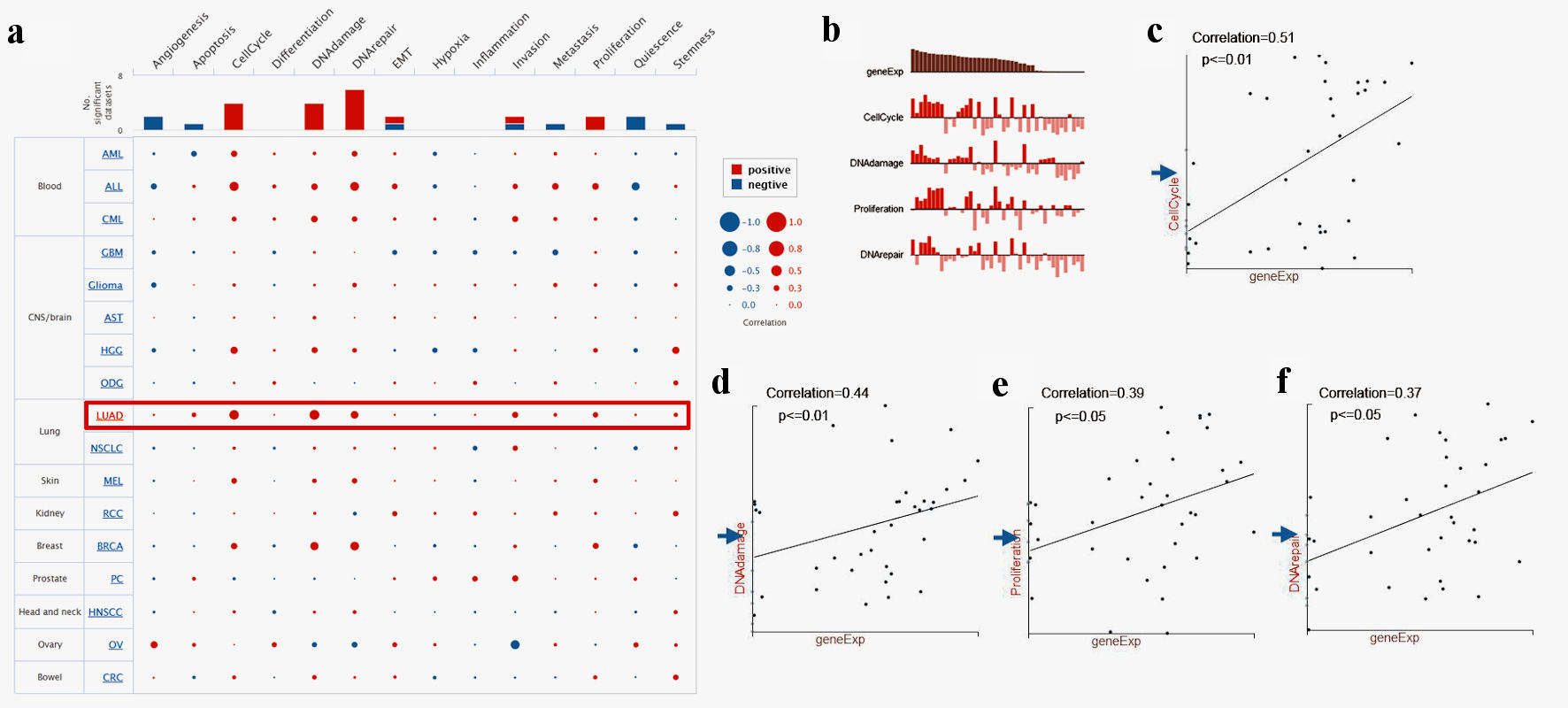 Click for large image | Figure 6. Single-cell functional analysis of TFDP1 in LUAD (CancerSEA). (a) Relevance of TFDP1 across 14 functions in multiple human cancers. (b-f) The various functions were markedly associated with TFDP1 in LUAD. TFDP1: transcription factor Dp-1; LUAD: lung adenocarcinoma. |
GSEA analysis of TFDP1 coexpressed genes in LUAD
To understand the genes related to TFDP1 and the mechanisms of TFDP1, LinkedOmics was employed to study TFDP1 mRNA sequencing data in 515 LUAD patients from the TCGA cohort. Figure 7a showed that 2,946 genes (dark red dots) were positively related to TFDP1, and 2,553 genes (dark green dots) were negatively related to TFDP1 in LUAD (FDR < 0.01). The top 50 positively TFDP1-correlated genes were demonstrated with heat maps in red (Fig. 7b) and the 50 negative genes in blue (Fig. 7c). Interestingly, survival maps of most positively and negatively TFDP1-correlated genes displayed high and low HRs, respectively (Fig. 7d). Moreover, TFDP1 expression was most positively correlated with CUL4A (positive rank# 1, Pearson correlation = 0.65, P = 3.703 × 10-63), TUBGCP3 (correlation = 0.6442, P = 1.057 × 10-61), PCID2 (correlation = 0.6172, P = 2.206 × 10-55), C13orf34 (correlation = 0.6136, P = 1.35 × 10-54), and C13orf37 (correlation = 0.5714, P = 5.786 × 10-46) (Fig. 7e), whereas TFDP1 was most negatively correlated with LMF1 (negative rank #1, correlation = -0.3745, P = 1.35 × 10-18), PBXIP1 (correlation = -0.3672, P = 7.019 × 10-18), COL4A3 (correlation = -0.3525, P = 1.62 × 10-16), C1orf116 (correlation = -0.3521, P = 1.79 × 10-16), and ZBTB4 (correlation = -0.3495, P = 3.08 × 10-16) (Fig. 7f).
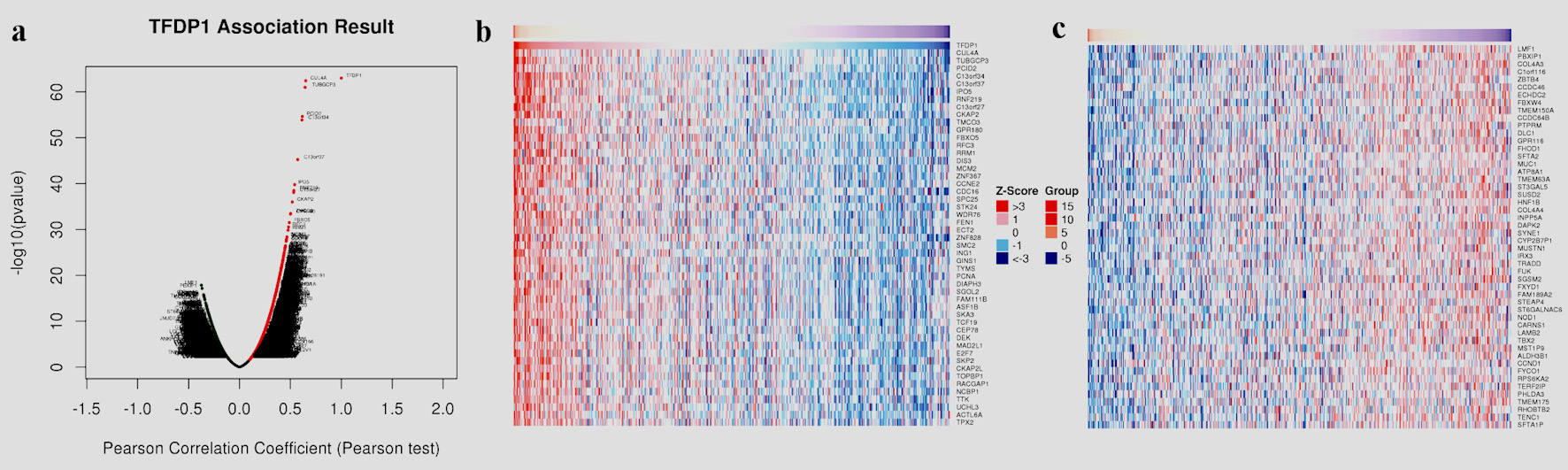 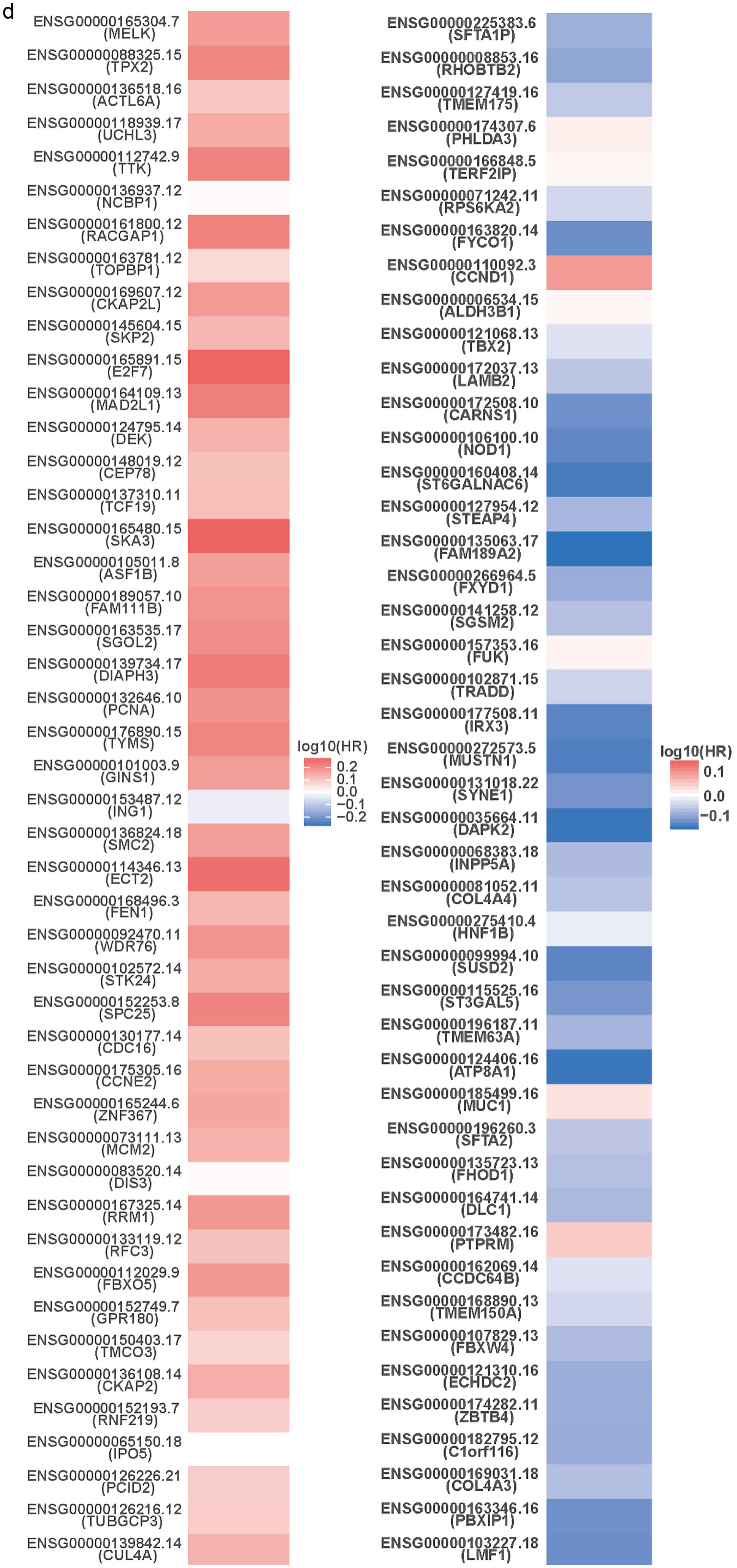 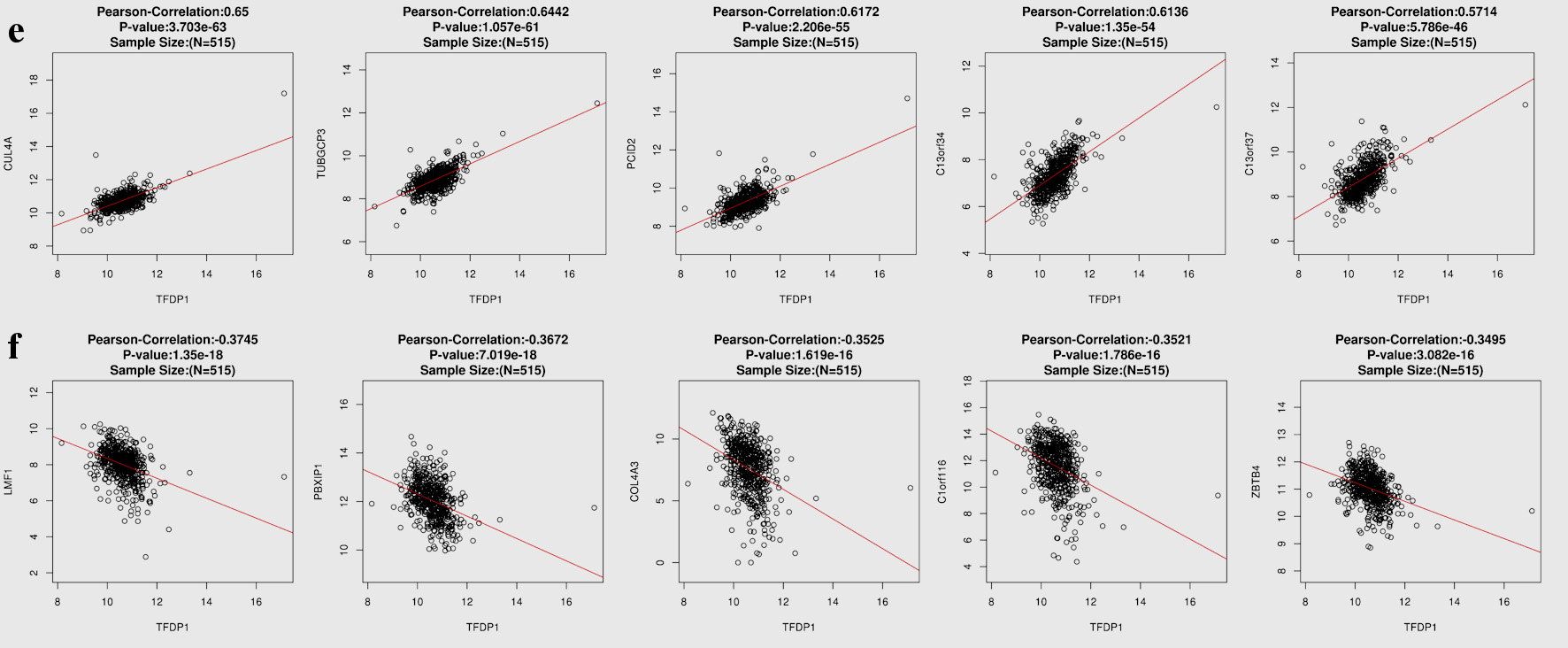 Click for large image | Figure 7. TFDP1 coexpressed genes in LUAD (LinkedOmics). (a) The volcano plot shows the TFDP1-related significantly coexpressed genes (Pearson test) in LUAD. (b) The heat map shows the top 50 positively TFDP1-correlated genes. (c) The heat map shows the top 50 negatively TFDP1-correlated genes. (d) Survival maps show the top 50 genes positively and negatively related to TFDP1, respectively. (e) Correlations between the top five positively TFDP1-correlated genes and TFDP1. (f) Correlations between the top five negatively TFDP1-correlated genes and TFDP1. TFDP1: transcription factor Dp-1; LUAD: lung adenocarcinoma. |
As shown in Figure 8, GO results of GSEA revealed that genes coexpressed with TFDP1 were mainly involved in the clusters related to cell proliferation, including chromosome segregation (Fig. 8c), DNA replication (Fig. 8d), spindle organization (Fig. 8e), cell cycle checkpoint (Fig. 8f), chromatin remodeling, telomere organization, and DNA recombination. KEGG pathway results also displayed that these genes were primarily enriched in cell proliferation, for example, cell cycle (Fig. 8h), DNA replication (Fig. 8i), spliceosome (Fig. 8k), homologous recombination (Fig. 8l), and nucleotide excision repair.
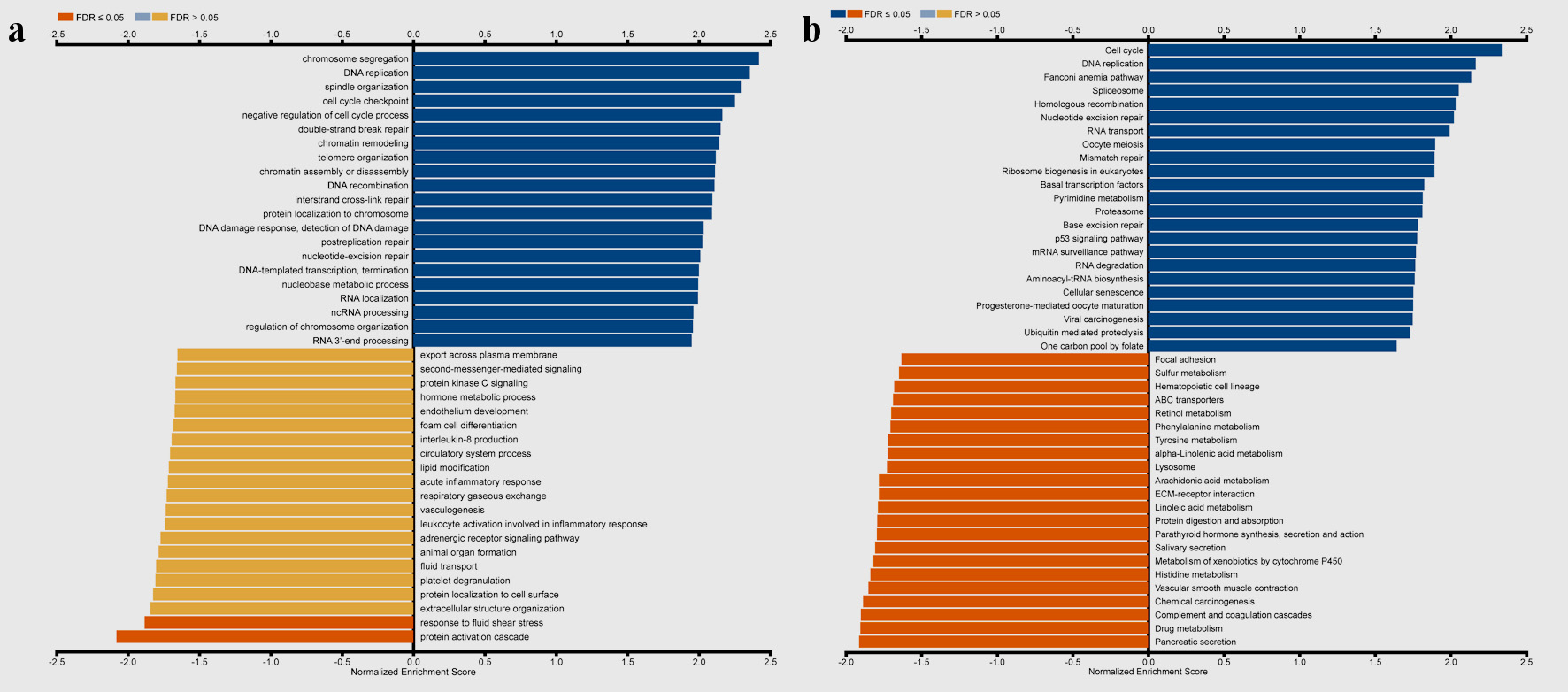 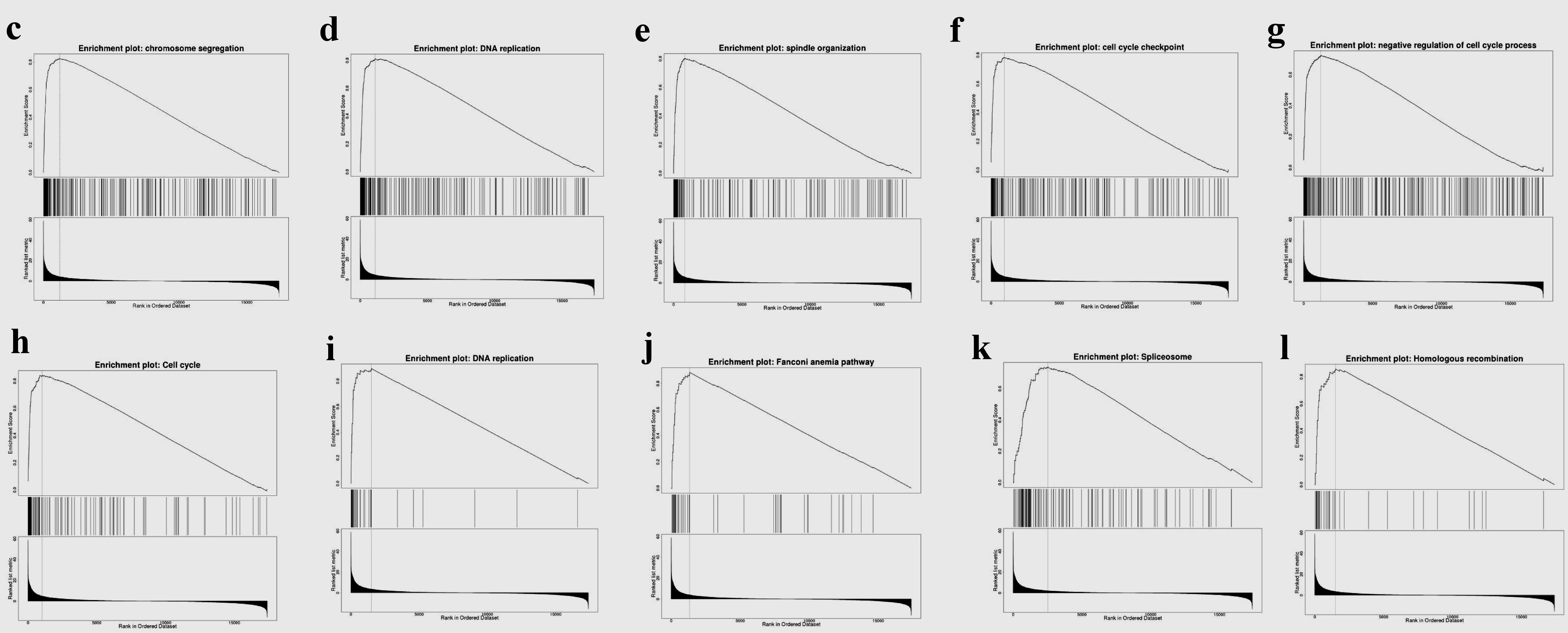 Click for large image | Figure 8. Gene set enrichment analysis of the genes coexpressed with TFDP1 in LUAD (LinkedOmics). (a) Biological process and (b) KEGG pathway analyses of TFDP1 in the TCGA cohort. (c) Chromosome segregation (normalized enrichment score (NES) = 2.4230, P < 0.001). (d) DNA replication (NES = 2.3660, P < 0.001). (e) Spindle organization (NES = 2.3299, P < 0.001). (f) Cell cycle checkpoint (NES = 2.2625, P < 0.001). (g) Negative regulation of cell cycle process (NES = 2.2039, P < 0.001). (h) Cell cycle (NES = 2.3389, P < 0.001). (i) DNA replication (NES = 2.1674, P < 0.001). (j) Fanconi anemia pathway (NES = 2.1372, P < 0.001). (k) Spliceosome (NES = 2.0545, P < 0.001). (l) Homologous recombination (NES = 2.0336, P < 0.001). TFDP1: transcription factor Dp-1; LUAD: lung adenocarcinoma KEGG: Kyoto Encyclopedia of Genes and Genomes; FDR: false discovery rate. |
Drug sensitivity analysis
We next used GSCA to analyze the drug sensitivity of mRNA levels of TFDP1 expression in the CTRP and GDSC databases. TFDP1 expression was positively related to the half-maximal inhibitory concentration (IC50) values of CHIR-99021, trametinib, bleomycin (50 µM), BEZ235, selumetinib, elesclomol, FH535, 17-AAG, docetaxel, and AG-014699 in the top 30 drugs from the GDSC database (Fig. 9a). Additionally, the details of more TFDP1-drug pairs can be found here (Supplementary Material 1, www.wjon.org). Figure 9b illustrated that the IC50 values of the top 30 small molecules were strongly negatively associated with TFDP1 mRNA expression in the CTRP database. Furthermore, the scores of the TFDP1-drug pairs showed that TFDP1 expression was positively associated with VAF-347 (cor = 0.125, FDR = 0.032) and SGX-523 (cor = 0.118, FDR = 0.02) (Supplementary Material 2, www.wjon.org). These results could provide the base for the TFDP1 potential roles in LUAD.
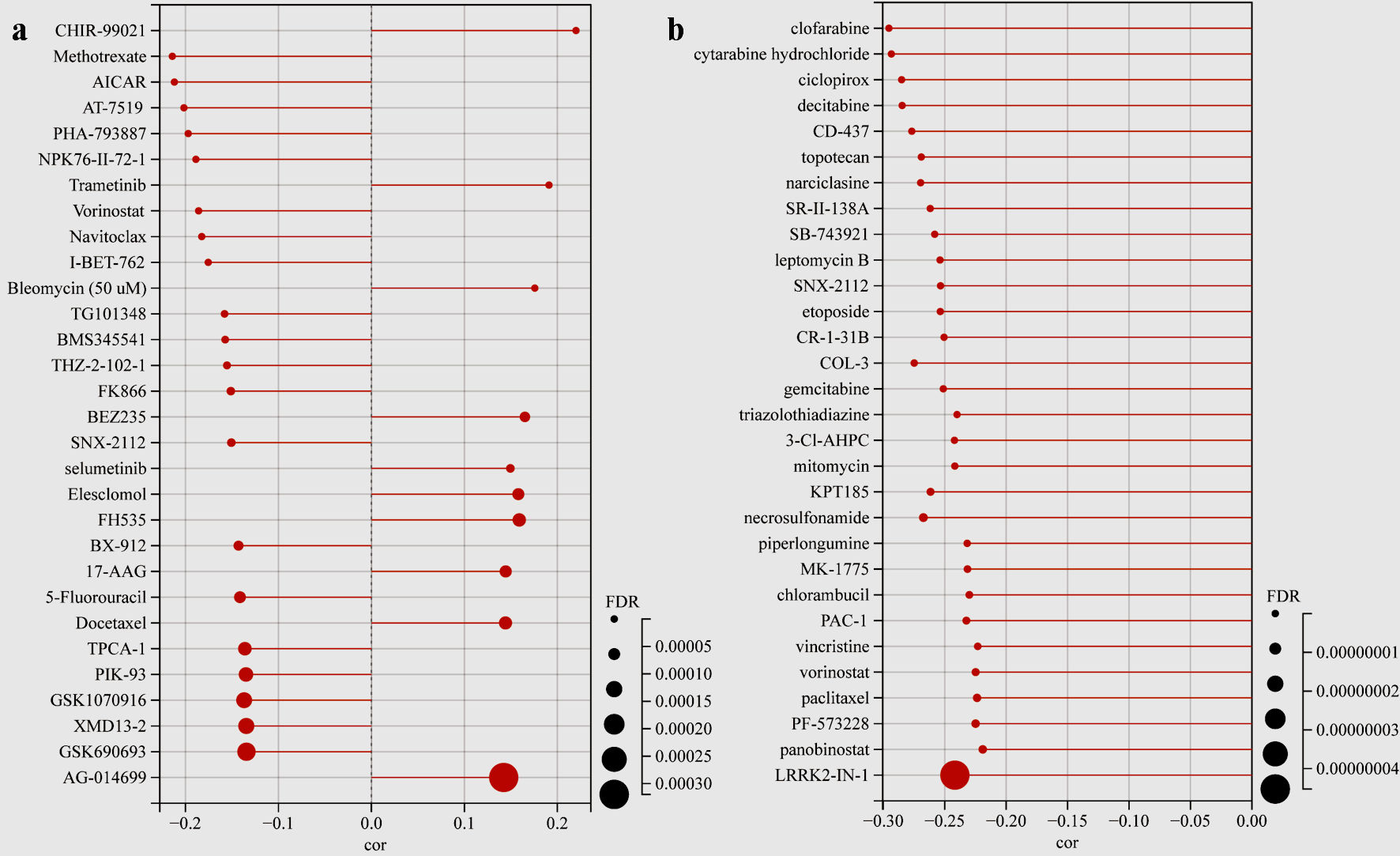 Click for large image | Figure 9. Drug sensitivity analysis. (a) The lollipop plot showing the TFDP1-drug pairs sensitivity in the GDSC database. (b) The lollipop plot showing the TFDP1-drug pairs sensitivity in the CTRP database. TFDP1: transcription factor Dp-1; GDSC: Genomics of Drug Sensitivity in Cancer; CTRP: Cancer Therapeutics Response Portal. |
| Discussion | ▴Top |
In this study, we first explored TFDP1 levels in different tissues from a pan-cancer perspective. Our results revealed that TFDP1 was markedly upregulated in three-fourths of 20 cancer types in the ONCOMINE data. The significant TFDP1 expression differences between tumor tissues and adjacent normal tissues for the lung (LUAD and lung squamous cell carcinoma (LUSC)) were confirmed from the TCGA dataset analyzed by TIMER. Additionally, the TFDP1 protein expression was significantly upregulated in LUAD tissues. Based on the Open Targets Platform, TFDP1 was related to various human diseases, including respiratory (LUAD and LUSC), thoracic, reproductive system, gastrointestinal, immune system, and endocrine system diseases. TFDP1 was overexpressed in breast and bladder cancer, confirmed by quantitative real-time polymerase chain reaction (qRT-PCR) and could be a potential biomarker [43, 44]. Jibrim et al used real-time polymerase chain reaction (PCR) to evaluate TFDP1 expression levels and found that TFDP1 expression was underexpressed in women’s endometrium [45]. From the UALCAN and GEPIA 2 databases, consistent results displayed that TFDP1 expression was strongly related to the LUAD cancer stages. As cancer stages advanced, TFDP1 mRNA expression levels elevated, and the highest levels were expressed in the highest cancer stage. Moreover, TFDP1 was overexpressed in all LUAD histological subtypes compared with normal tissues and significantly upregulated in several subtypes. We also found that AJCC pathologic stage, AJCC stage T, and AJCC stage N were the independent prognostic factors in LUAD. High expression of TFDP1 might mean poor OS, FPS, and PPS valus in LUAD patients. These findings unlocked that TFDP1 was overexpressed and could be a potential prognostic biomarker in LUAD. Abba et al showed that TFDP1 could be a target of overexpression and amplification in breast cancer [46]. Barh et al revealed that TFDP1 was overexpressed in the NSCLC stage-II and IV blood and could be a potential diagnostic marker in NSCLC [47], which supported our findings. Zhang et al revealed that high TFDP1 expression implied a poor prognosis in oral squamous cell carcinoma [48]. Yang et al identified that TFDP1 expression might be related to significant somatic copy number aberrations in Chinese papillary THCA [49]. All these studies showed that TFDP1 expression levels were abnormally expressed in various cancer tissues and played a crucial role in multiple cancer types.
Furthermore, cBioPortal was utilized to explore TFDP1 genomic alteration of LUAD patients. The results showed that deep deletion (10 cases), mutation (nine cases), amplification (two cases), and multiple alterations (one case) of TFDP1 were found in 566 LUAD patients. Moreover, LUAD patients with TFDP1 alterations were associated with poor OS and DFS. Combined with our above findings, TFDP1 might be involved in all stages of LUAD progression and could be used as a potential target of LUAD treatment. However, the development and progression of cancers were usually promoted by the combined action of multiple cooccurrences of gene alterations [11, 13, 16]. Therefore, we found that RYR2, MUC16, TTN, ZFHX4, TP53, GRK1, LAMP1, RASA3, CSMD3, and ATP4B cooccurred with TFDP1 alterations and alteration patterns of TFDP1 cooccurred with GRK1, LAMP1, RASA3, and ATP4B in LUAD patients. Then, we constructed a gene-gene interaction network of the five cooccurred genes. Wang et al found that Rac3 acted on TFDP1, CCND1, and MYC to regulate cell proliferation of LUAD during tumorigenesis [50]. Ren et al found that RYR2 mutation could lead to a better prognosis by downregulation of DKK1, which was associated with TP53, MTOR, and VEGF in NSCLC [51]. Therefore, the specific mechanism of genes cooccurred with TFDP1 alterations in LUAD may be worth further research.
DNA methylation is a key epigenetic factor for various cancer types and has been employed to diagnose different cancers [52-54]. Inoshita et al found that TFDP1 was a distinctive sex-biased DNA methylation gene in peripheral blood samples [55]. DNA methylation levels of TFDP1 were high in human islets [56]. Next, we used MethSurv to find LUAD patients with hypermethylation of CpG cg26355737 and cg01595804 led to a good prognosis. Liu et al revealed that low TFDP1 DNA methylation levels were strongly related to poor prognosis in LUAD [57], which strongly supported our research.
We used LinkedOmics to reveal that higher TFDP1 levels were markedly related to the worse prognosis, and most of the genes coexpressed with TFDP1 in LUAD were also strongly correlated with the prognosis. Liu et al found that TFDP1 and CUL4A were poor prognosticators of intrahepatic cholangiocarcinoma [58], which might agree with our research. Moreover, GO and KEGG pathway results revealed that TFDP1 mainly participated in multiple biological processes related to cell proliferation (such as DNA replication, double-strand break repair, cell cycle checkpoint, and protein activation cascade) and KEGG pathways of proliferation (such as spliceosome, DNA replication, cell cycle, and homologous recombination). Tavana et al found that TFDP1 could impact on the cell cycle of cattle follicles [59]. Zhan et al revealed that COMMD9 interacted with TFDP1 to enhance the p53 signaling pathway in NSCLC [16]. Zhang et al found that TFDP1 participated in PIP4K2A transcriptional regulatory network and influenced the survival rate of various cancers [60]. Jiang et al found that MycoPhyto® Complex inhibited TFDP1 expression to regulate breast cancer cell cycle [61]. Yang et al. found that TFDP1 was one of the driver genes participating in several significant driver pathways of Chinese papillary THCA [49]. These studies showed that TFDP1 was a key to the cell cycle and involved in several pathways of different human cancers, which strongly supported our findings.
Interestingly, to further verify our research, we used CancerSEA to reveal that TFDP1 was markedly related to cell cycle, DNA damage, proliferation, and DNA repair. Recent research showed that Rac3 regulated LUAD cell proliferation by acting on TFDP1, CCND1, and MYC related to the cell cycle pathway during tumorigenesis [50]. Bisikirska et al showed that TFDP1 could promote the progression of follicular lymphoma and was a candidate regulator [62]. Therefore, our findings indicated that TFDP1 and its coexpressed genes could act on the LUAD development and progression by the combined action on the cell cycle process. KEGG pathway results revealed that genes coexpressed with TFDP1 were enriched in drug metabolism (LinkedOmics), and chemotherapy patients with TFDP1 high mRNA levels tended to have poor OS in LUAD (HR = 11.57, 95% CI: 2.22 - 60.26), P = 0.00069) (Kaplan-Meier plotter). Therefore, the potential roles of TFDP1-coexpressed genes in therapeutic responses of LUAD patients need further research.
Finally, the drug sensitivity of mRNA levels of TFDP1 expression in the CTRP and GDSC databases was analyzed by GSCA. The GDSC lollipop plot showed that TFDP1 expression was positively related to CHIR-99021, trametinib, bleomycin (50 µM), BEZ235, selumetinib, elesclomol, FH535, 17-AAG, docetaxel, and AG-014699. Although TFDP1 mRNA expression levels were strongly negatively related to the IC50 values of all the top 30 CTRP small molecules, it is revealed that TFDP1 expression was positively associated with VAF-347 and SGX-523 (Supplementary Material 2, www.wjon.org). Thus, further research could be needed to verify these results before clinical practices in LUAD.
Conclusions
In summary, we comprehensively analyzed expression levels, clinicopathological features, prognosis, genomic alterations, DNA methylation, functional states, related genes, functional enrichment, and drug sensitivity of TFDP1 in LUAD patients. Our results unveiled that TFDP1 was obviously upregulated in most human cancers and related to various diseases, including LUAD. TFDP1 was markedly related to individual cancer stages, and high TFDP1 expression in LUAD might imply a shorter prognostic value. Moreover, TFDP1 alterations resulted in poor OS and DFS, and LUAD patients with hypermethylation might have a good prognosis. Genomic alterations and DNA methylation of TFDP1 might impact the prognosis of LUAD patients. TFDP1 and its coexpressed genes might affect the LUAD development and progression by the combined action on the cell cycle process. TFDP1 mainly participated in multiple signaling pathways of cell proliferation, such as the cell cycle, DNA replication, and spliceosome. Then, we found that TFDP1 was strongly positively related to the IC50 values of several drugs. Our findings showed that TFDP1 might be a potential prognostic biomarker for LUAD and provide the base for potential targeted agents in LUAD patients.
| Supplementary Material | ▴Top |
Suppl 1. The scores of the TFDP1-drug pairs in the GDSC database.
Suppl 2. The scores of the TFDP1-drug pairs in the CTRP database.
Acknowledgments
None to declare.
Financial Disclosure
This study is supported by the National Social Science Fund of China (18CFX059).
Conflict of Interest
The authors declare that they have no conflict of interest.
Informed Consent
Not applicable.
Author Contributions
YS: conceptualization, data curation, investigation, methodology, visualization, writing of original manuscript, and final manuscrip. RM: conceptualization, formal analysis, methodology, supervision, writing of final manuscript. All authors have read and agreed to the published version of the manuscript.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
| References | ▴Top |
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249.
doi pubmed - Cao M, Li H, Sun D, Chen W. Cancer burden of major cancers in China: A need for sustainable actions. Cancer Commun (Lond). 2020;40(5):205-210.
doi pubmed pmc - Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543-550.
doi pubmed pmc - Brustugun OT, Gronberg BH, Fjellbirkeland L, Helbekkmo N, Aanerud M, Grimsrud TK, Helland A, et al. Substantial nation-wide improvement in lung cancer relative survival in Norway from 2000 to 2016. Lung Cancer. 2018;122:138-145.
doi pubmed - Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr., Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299-311.
doi pubmed - Jurisic V, Vukovic V, Obradovic J, Gulyaeva LF, Kushlinskii NE, Djordjevic N. EGFR polymorphism and survival of NSCLC patients treated with TKIs: a systematic review and meta-analysis. J Oncol. 2020;2020:1973241.
doi pubmed pmc - Girling R, Partridge JF, Bandara LR, Burden N, Totty NF, Hsuan JJ, La Thangue NB. A new component of the transcription factor DRTF1/E2F. Nature. 1993;362(6415):83-87.
doi pubmed - Bandara LR, Lam EW, Sorensen TS, Zamanian M, Girling R, La Thangue NB. DP-1: a cell cycle-regulated and phosphorylated component of transcription factor DRTF1/E2F which is functionally important for recognition by pRb and the adenovirus E4 orf 6/7 protein. EMBO J. 1994;13(13):3104-3114.
doi pubmed pmc - Zaragoza K, Begay V, Schuetz A, Heinemann U, Leutz A. Repression of transcriptional activity of C/EBPalpha by E2F-dimerization partner complexes. Mol Cell Biol. 2010;30(9):2293-2304.
doi pubmed pmc - Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, Palacios J, et al. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009;11(6):R86.
doi pubmed pmc - Pellicelli M, Picard C, Wang D, Lavigne P, Moreau A. E2F1 and TFDP1 Regulate PITX1 expression in normal and osteoarthritic articular chondrocytes. PLoS One. 2016;11(11):e0165951.
doi pubmed pmc - Chen C, Liu J, Zhou F, Sun J, Li L, Jin C, Shao J, et al. Next-generation sequencing of colorectal cancers in chinese: identification of a recurrent frame-shift and gain-of-function Indel mutation in the TFDP1 gene. OMICS. 2014;18(10):625-635.
doi pubmed pmc - Morimoto Y, Mizushima T, Wu X, Okuzaki D, Yokoyama Y, Inoue A, Hata T, et al. miR-4711-5p regulates cancer stemness and cell cycle progression via KLF5, MDM2 and TFDP1 in colon cancer cells. Br J Cancer. 2020;122(7):1037-1049.
doi pubmed pmc - Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, Emi M, et al. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35(6):1476-1484.
doi pubmed - Drucker E, Holzer K, Pusch S, Winkler J, Calvisi DF, Eiteneuer E, Herpel E, et al. Karyopherin alpha2-dependent import of E2F1 and TFDP1 maintains protumorigenic stathmin expression in liver cancer. Cell Commun Signal. 2019;17(1):159.
doi pubmed pmc - Zhan W, Wang W, Han T, Xie C, Zhang T, Gan M, Wang JB. COMMD9 promotes TFDP1/E2F1 transcriptional activity via interaction with TFDP1 in non-small cell lung cancer. Cell Signal. 2017;30:59-66.
doi pubmed - Wang Z, Embaye KS, Yang Q, Qin L, Zhang C, Liu L, Zhan X, et al. Development and validation of a novel epigenetic-related prognostic signature and candidate drugs for patients with lung adenocarcinoma. Aging (Albany NY). 2021;13(14):18701-18717.
doi pubmed pmc - NGS. https://www.oncomine.org/.
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1-6.
doi pubmed pmc - TIMER. https://cistrome.shinyapps.io/timer/.
- Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108-e110.
doi pubmed pmc - UALCAN. http://ualcan.path.uab.edu/.
- Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649-658.
doi pubmed pmc - Open Targets Platform. https://www.targetvalidation.org/.
- Carvalho-Silva D, Pierleoni A, Pignatelli M, Ong C, Fumis L, Karamanis N, Carmona M, et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 2019;47(D1):D1056-D1065.
doi pubmed pmc - GEPIA2. http://gepia2.cancer-pku.cn/.
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556-W560.
doi pubmed pmc - Kaplan-Meier Plotter. http://kmplot.com/analysis/.
- Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241.
doi pubmed pmc - cBioPortal. http://www.cbioportal.org/.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-404.
doi pubmed pmc - Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.
doi pubmed pmc - GENEMANIA. http://www.genemania.org/.
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214-W220.
doi pubmed pmc - MethSurv. https://biit.cs.ut.ee/methsurv/.
- Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277-288.
doi pubmed - CancerSEA. http://biocc.hrbmu.edu.cn/CancerSEA/.
- Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47(D1):D900-D908.
doi pubmed pmc - LinkedOmics. http://www.linkedomics.org/.
- Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956-D963.
doi pubmed pmc - GSCA. http://bioinfo.life.hust.edu.cn/GSCA/.
- Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771-3772.
doi pubmed - Moscovich M, LeDoux MS, Xiao J, Rampon GL, Vemula SR, Rodriguez RL, Foote KD, et al. Dystonia, facial dysmorphism, intellectual disability and breast cancer associated with a chromosome 13q34 duplication and overexpression of TFDP1: case report. BMC Med Genet. 2013;14:70.
doi pubmed pmc - Alekseev B, Vorobyev N, Shegay P, Zabolotneva A, Rusakov I, Buzdin A, Gaifullin N, et al. Mp22-18 identification of novel gene expression markers for bladder cancer diagnostics. The Journal of Urology. 2014;191(4S):e241-e242.
- Jibrim RLM, de Carvalho CV, Invitti AL, Schor E. Expression of the TFDP1 gene in the endometrium of women with deep infiltrating endometriosis. Gynecol Endocrinol. 2019;35(6):490-493.
doi pubmed - Abba MC, Fabris VT, Hu Y, Kittrell FS, Cai WW, Donehower LA, Sahin A, et al. Identification of novel amplification gene targets in mouse and human breast cancer at a syntenic cluster mapping to mouse ch8A1 and human ch13q34. Cancer Res. 2007;67(9):4104-4112.
doi pubmed pmc - Barh D, Jain N, Tiwari S, Field JK, Padin-Iruegas E, Ruibal A, Lopez R, et al. A novel in silico reverse-transcriptomics-based identification and blood-based validation of a panel of sub-type specific biomarkers in lung cancer. BMC Genomics. 2013;14(Suppl 6):S5.
doi pubmed pmc - Zhang L, Li H, Qiu Y, Liu Y, Liu X, Wang W. Screening and cellular validation of prognostic genes regulated by super enhancers in oral squamous cell carcinoma. Bioengineered. 2021;12(2):10073-10088.
doi pubmed pmc - Yang C, Xu W, Gong J, Liu Z, Cui D. Novel somatic alterations underlie Chinese papillary thyroid carcinoma. Cancer Biomark. 2020;27(4):445-460.
doi pubmed - Wang G, Wang H, Zhang C, Liu T, Li Q, Lin X, Xie J, et al. Rac3 regulates cell proliferation through cell cycle pathway and predicts prognosis in lung adenocarcinoma. Tumour Biol. 2016;37(9):12597-12607.
doi pubmed - Ren W, Li Y, Chen X, Hu S, Cheng W, Cao Y, Gao J, et al. RYR2 mutation in non-small cell lung cancer prolongs survival via down-regulation of DKK1 and up-regulation of GS1-115G20.1: A weighted gene Co-expression network analysis and risk prognostic models. IET Syst Biol. 2022;16(2):43-58.
doi pubmed pmc - Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21(35):5450-5461.
doi pubmed - Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457-466.
doi pubmed - Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012;33(2):287-296.
doi pubmed - Inoshita M, Numata S, Tajima A, Kinoshita M, Umehara H, Yamamori H, Hashimoto R, et al. Sex differences of leukocytes DNA methylation adjusted for estimated cellular proportions. Biol Sex Differ. 2015;6:11.
doi pubmed pmc - Hall E, Volkov P, Dayeh T, Esguerra JL, Salo S, Eliasson L, Ronn T, et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 2014;15(12):522.
doi pubmed pmc - Liu H, Zhao H. Prognosis related miRNAs, DNA methylation, and epigenetic interactions in lung adenocarcinoma. Neoplasma. 2019;66(3):487-493.
doi pubmed - Liu TT, You HL, Weng SW, Wei YC, Eng HL, Huang WT. Recurrent amplification at 13q34 targets at CUL4A, IRS2, and TFDP1 as an independent adverse prognosticator in intrahepatic cholangiocarcinoma. PLoS One. 2015;10(12):e0145388.
doi pubmed pmc - Tavana A, Farhangfar H, Behdani E. Fitting interaction network among effective transcription factors associated with ovulation rate and explored genes by using promoter analysis in cattle. Journal of Animal Science Research. 2021;31(2):45-56.
- Zhang S, Li Z, Yan X, Bao L, Deng Y, Zeng F, Wang P, et al. Regulatory network and prognostic effect investigation of PIP4K2A in leukemia and solid cancers. Front Genet. 2018;9:721.
doi pubmed pmc - Jiang J, Sliva D. Novel medicinal mushroom blend suppresses growth and invasiveness of human breast cancer cells. Int J Oncol. 2010;37(6):1529-1536.
doi pubmed - Bisikirska B, Bansal M, Shen Y, Teruya-Feldstein J, Chaganti R, Califano A. Elucidation and pharmacological targeting of novel molecular drivers of follicular lymphoma progression. Cancer Res. 2016;76(3):664-674.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


