| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website http://www.wjon.org |
Original Article
Volume 8, Number 5, October 2017, pages 162-170
CTLA-4 Genetic Variants (rs11571317 and rs3087243): Role in Susceptibility and Progression of Breast Cancer
Maruthi Goskea, V. R. Vinish Ramachandera, Prasanna Latha Komaravallia, P. Fazul Rahmanb, Chandrasekhar Raoc, Parveen Jahanb, d
aDepartment of Genetics, Osmania University, Hyderabad-07, Telangana, India
bDepartment of Zoology, Maulana Azad National Urdu University, Gachibowli, Hyderabad-32, Telangana, India
cSoumya Multi Specialty Hospital, Hyderabad-07, Telangana, India
dCorresponding Author: Parveen Jahan, Department of Zoology, Maulana Azad National Urdu University, Hyderabad 500032, Telangana, India
Manuscript submitted June 13, 2017, accepted August 21, 2017
Short title: CTLA-4 Genetic Variants
doi: https://doi.org/10.14740/wjon1046w
| Abstract | ▴Top |
Background: Dysfunctional regulation at immune checkpoints may lead to escape of the tumor cells and gives a scope to set in the unresolved Breast cancer (BC). The major anti-tumor retort is cell-mediated response which involves T lymphocytes. CTLA-4 (Cytotoxic T lymphocyte associated protein-4) with immune suppressive function and tolerance is associated with various autoimmune diseases and cancers including BC. The present study deals with CTLA-4 gene selected polymorphisms (rs11571317 C/T and rs3087243G/A) to explore their relation with breast cancer susceptibility and progression in BC patients.
Methods: For the present case-control study, we recruited a total of 570 women which include breast cancer patients and healthy control women from south India. Blood samples were collected, genomic DNA was isolated and genotyped by using PCR-RFLP method, and the data were analysed through suitable statistics.
Results: We observed a significant association of rs11571317 with BC in our study group, where CC genotype showed a three-fold increased risk towards BC and CT genotype to be protective. In-silico analyses strengthened our observation revealing the abolition of SP1 binding site in the CTLA-4 promoter by the mutant allele T. The CTLA-4 rs3087243 polymorphism showed an association not with the susceptibility but towards the tumor progression, where GG genotype was coupled with reduced tumor growth (OR = 0.01) and GA (OR = 6.2), AA (OR = 3.4) with increased tumor growth. The T-G haplotype was found to confer protection against breast cancer risk while C-A (OR = 3.6) and T-A (OR = 15.8) haplotypes were associated with disease progression. In-silico analysis for rs3087243 revealed change in threshold values between reference and variant sequences.
Conclusion: The study suggests varied roles of different polymorphisms of CTLA-4 in the aetiopathogenesis of BC. Understanding the mechanism may help in the CTLA-4 based immunotherapy for BC.
Keywords: Breast Cancer; Immune regulatory; T-cells; CTLA-4
| Introduction | ▴Top |
Breast cancer (BC) is the most common malignancy threatening the health and life of women where its incidence has increased in recent years in both developed and developing countries [1-3]. According to the data of International Agency for Research on Cancer, there were 1.68 million new breast cases reported in recent years. BC is characterized as non-cutaneous malignancy associated with uncontrolled growth in ducts (ductal carcinoma) or lobules (lobular carcinoma) of the mammary gland [4]. These tumors can invade the surrounding tissues of the breast in later stages. The major influences on BC appear to be environmental and genetic factors [5]. Previous studies indicated that genetic influence accounts for about 27% of the BC risks [6].
BC is an extremely complex disease; its onset and progression is a multi-step process resulting from a series of epigenetic, genetic, endocrine and external environmental factors [7, 8]. The role of genetic factors in epidemiology and pathogenesis of both sporadic (genetic mutations) and familial BCs (inherited defects in DNA repair genes BRCA1 and BRCA2) and failure of immune surveillance (low cell-mediated immunity) is now well established [9-12].
Cytotoxic T-lymphocyte associated protein-4 (CTLA-4), a key immune regulatory molecule, is involved in tolerance and down-regulation of T-cell activation. CTLA-4 protein structure contains an extracellular V domain, a transmembrane domain, and a cytoplasmic tail. Alternate splice variants of different CTLA-4-encoding isoforms have been characterized. The membrane-bound isoform functions as a homodimer and is interconnected by a disulfide bond, while the soluble isoform functions as a monomer. The gene for CTLA-4 is located at 2q33 and several single nucleotide polymorphisms (SNPs) in this gene are known to have an effect on its genetic expression and protein activity [13]. Previous studies have extensively examined the association of polymorphisms within CTLA-4 with autoimmune diseases including Graves’ disease, hepatocellular carcinoma, breast carcinoma, renal disease, type I diabetes, systemic lupus erythematosus and vitiligo [14-20]. However, we did not find any studies in the context of CTLA-4 gene polymorphism in BC from south Indian population.
The two selected polymorphic sites of the CTLA-4 gene for the present study were -658 T/C (rs11571317) and CT60 G/A (rs3087243). A study on -658 T/C polymorphism had been shown to be associated with BC in Chinese Han population (2007); however, the functional significance of this polymorphism is not clear. On the other hand, SNP CT60 G/A present in the 3'-UTR of CTLA-4 gene was described as a complex region containing multiple binding sites for microRNA, which is suggested to influence the signaling pathway and protein expression that may result in diseases [14, 21].
| Materials and Methods | ▴Top |
Subjects
Blood samples were collected from a total of 570 individuals comprising 285 BC patients and 285 healthy controls. The patients were recruited from the Osmania General Hospital and Railway Central Hospital, Soumya Multi-specialty Hospital and ESI Hospital, Hyderabad, India. All patients were either histopathologically or cytologically confirmed as having BC. Clinical and demographic information was obtained from medical reports and through personal interaction. The controls were healthy volunteers who are age-matched women visiting general health checkup division. All the subjects enrolled in the study were unrelated residents of south India. The study has been approved by institutional ethical committee for Biomedical Research, Osmania University, Hyderabad, India, in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all study participants at the time of sample collection.
Genotyping for CTLA-4 gene-selected polymorphisms: rs11571317 and rs3087243
Genomic DNA was isolated by salting out method using standard protocol in our lab [22]. Genotyping was done by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) method. The primers for PCR amplification of rs11571317 and rs3087243 were used as described [16]. PCR was performed in a final volume of 10-µL reaction mixture containing 2 µL of genomic DNA, 0.2 µmol/L of each primer, 0.2 µmol/L of each dNTP, 0.2 µL of Taq DNA polymerase (5 U Labpro India), and 10 × PCR buffer supplied by Labpro India. The PCR profile consisted of an initial melting step of 5 min at 94 °C, followed by 27 cycles of 30 s, secondary melting temperature at 94 °C, 45 s and annealing temperature at 52 °C, 55 s for rs11571317 and for rs3087243 annealing temperature at 51.6 °C, 50 s and a final elongation at 72 °C for 5 min. The restriction endonucleases Aci I for rs11571317 and HpyCH4 IV for rs3087243 SNP (New England Biolabs, UK) were used to distinguish the CTLA-4 genotypes. The amplified products were run on 2% agarose gel, containing ethidium bromide at 100 V for 20 min. Post-PCR amplification product of rs11571317 was 366 bp which was digested with Aci I restriction endonuclease that resulted in fragment sizes of 254 and 112 bp for CC, 366, 254, and 112 bp for CT and undigested product of 366 bp for TT genotype. Similarly when 178 bp product of rs3087243 was digested with HpyCH4 IV, the resultant fragment sizes of 107 and 72 bp (not seen on gel) for GG, 178 and 107 bp for GA and 178 bp for AA genotype were visualized under gel documentation system (UV Tech, UK), as shown in Figure 1.
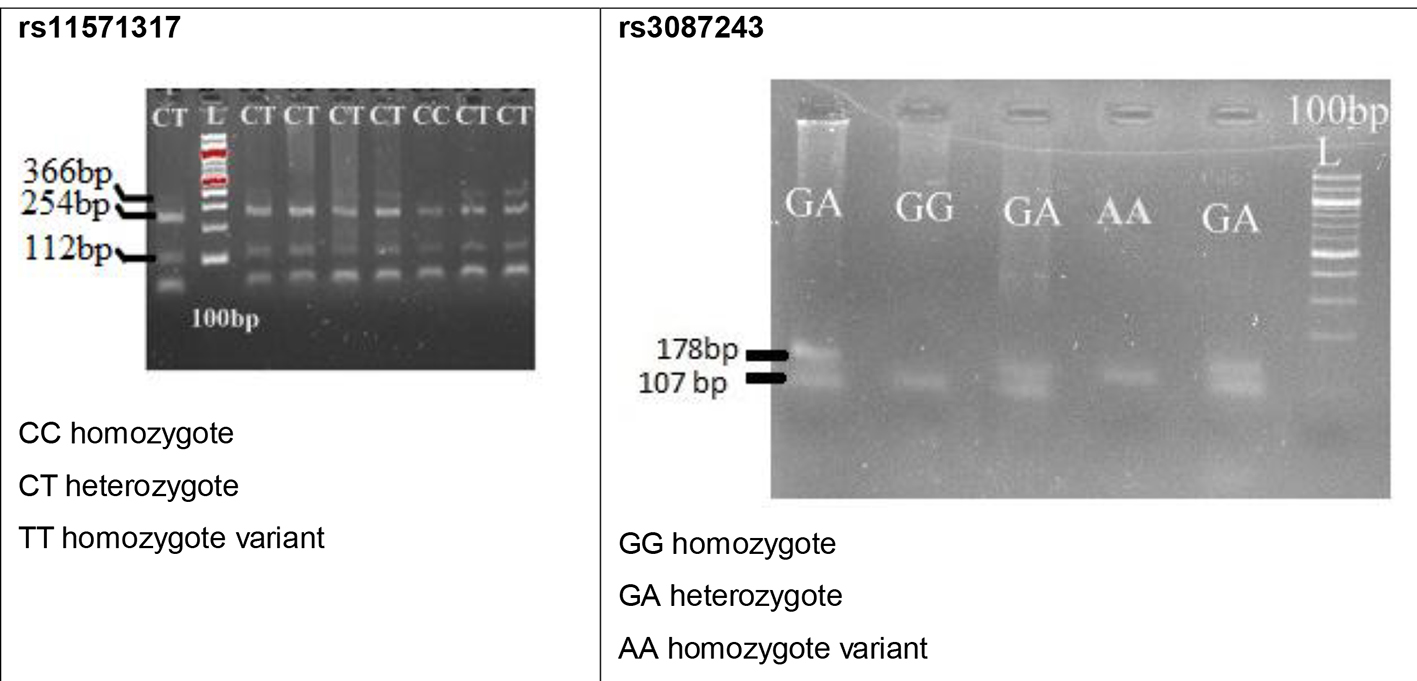 Click for large image | Figure 1. Gel images of selected polymorphisms in the present study. |
Statistical analysis
Descriptive statistics were used to calculate percentages, mean, and standard deviation. Z-test was carried out to calculate the difference in the proportions of epidemiological variables. Mean differences between patients and controls with respect to age and body mass index (BMI) were done by t-test. Chi-square test was used to compare the allele and genotype frequencies between patients and controls as well as within the subgroups. Hardy-Weinberg equilibrium (HWE) was tested using a calculator by Michael H Court for comparing the observed and expected genotype frequencies in cases and controls. The risk-associated genotypes were calculated using odds ratio (OR) at 95% confidence interval (CI) with two-tailed level of significance (< 0.05). Multiple logistic regression analysis was carried out for selected CTLA-4 SNPs with clinical parameters (http://statpages.info/logistic.html). Pairwise linkage disequilibrium (LD) analysis was done using Haploview software (version 4.2). Data analysis was performed by SPSS version 21. In-silico analysis was carried out using transcription factors prediction and splicing finder analysis tools.
In the present study, demographic and clinical aspects considered for analysis were age at onset of the disease (≤ 48 years vs. > 48 years), tumor status (T1-2 vs. T3-4), lymph node involvement (LN-ve vs. LN+ve), metastasis (M0 vs. M1) and duration of disease (DOD, < 6 months vs. > 6 months), as shown in Table 1.
 Click to view | Table 1. Clinical Characteristics of BC Patients |
| Results | ▴Top |
Demographic and clinical data
The demographic and clinical information about the cases and the controls enrolled in the present study is summarized in Tables 1 and 2. Significant variation was noted between patients and controls with respect to parity, consanguinity, BMI, breast feeding, area of living and occupation.
 Click to view | Table 2. Demographical and Clinical Information of Breast Cancer Patients and Healthy Controls |
Analysis of CTLA-4 rs11571317 polymorphism
The distributions of CC, CT and TT genotypes of CTLA-4 rs11571317 were 13.5%, 86.5% and 0% in controls and 31%, 69% and 0% in patients. A statistically significant association between genotypes of CTLA-4 rs11571317 and BC risk was observed (χ2 = 24.19, P = 0.01). OR revealed an increased frequency of homozygote CC genotype in patients compared to controls with an OR of 2.86 (CI = 1.80 - 4.36, P < 0.01); the heterozygous genotype CT figured prominently among the controls and revealed an OR of 0.34 (CI = 0.22 - 0.53; P < 0.01). No association was found for alleles C and T with BC risk, and genotypes were not consistent with HWE (controls χ2 = 164.2, patients χ2 = 78.2, P < 0.05).
MLR analysis of patients showed lack of association of rs11571317 genotypes with any of the parameters such as age at onset of the disease, tumor size, lymph node status, metastasis and DOD (P > 0.05), as shown in Table 3.
 Click to view | Table 3. Genotype and Allele Frequencies of CTLA-4 (rs11571317) Gene Polymorphism in South Indian Breast Cancer Patients, Healthy Controls and Multiple Logistic Regression Analysis for BC Clinical Pathological Features |
In-silico analysis
The in-silico analysis of CTLA-4 gene SNP rs11571317 by TF search tool alibaba 2.1 predicted the possible transcription factor association with the CTLA-4 gene polymorphisms in normal and polymorphic variant sequences (Fig. 2).
 Click for large image | Figure 2. The in-silico analysis showed CTLA-4 gene rs11571317 SNP and possible associated transcription factors. |
Analysis of CTLA-4 rs3087243 polymorphism
The distributions of GG, GA and AA genotypes of CTLA-4 rs3087243 were 26%, 71% and 3% in controls and 25%, 70% and 5% in patients and there was no association between genotypes and BC risk (χ2 = 1.95, P > 0.05). Similarly, no significant relation was found between alleles G and A with BC risk. Genotypes were not in consistent with HWE (controls χ2 = 69.9, patients χ2 = 53.6, P < 0.05).
MLR analysis showed that clinical parameter such as tumor status was significantly associated with rs3087243 polymorphism. The homozygote wild-type GG genotype was predominant in early tumor status, i.e. T1-2 (OR = 0.01, CI = 0.0043 - 0.09, P < 0.01), heterozygote GA in advanced tumor, i.e. T3-4 (OR = 6.28, CI = 3.01 - 13.07, P < 0.01), and AA genotype frequency in T3-4 (OR = 3.46, CI = 1.02 - 11.67, P < 0.01). However, there was no significant association with age at onset of disease, lymph node involvement, metastasis and DOD (P > 0.05), as shown in Table 4.
 Click to view | Table 4. Genotype and Allele Frequencies of CTLA-4 rs3087243 Gene Polymorphism in South Indian Breast Cancer Patients, Healthy Controls and Multiple Logistic Regression Analysis for BC Clinical Pathological Features |
In-silico analysis
We performed splicing finder tool to CTLA-4 SNP rs3087243 which was located in 3’UTR region of CTLA-4 gene. The results are mentioned in Table 5.
 Click to view | Table 5. The Reference Motif cgtgggtt (78.23) Threshold Value Is Changed to Mutant catgggtt (51.7) |
Haplotype analysis
Further, the four possible haplotypes C-G, C-A, T-G and T-A were demonstrated in our population (Table 6). The most frequent haplotype was observed to be T-G haplotype in both patients and controls with significant difference (harboring one mutant allele and one ancestral allele) (OR = 0.40, CI = 0.25 - 0.62, P = 0.01). In addition, tumor size, metastasis and DOD showed a significant association (P < 0.01) with C-A and T-A haplotypes. The C-A and T-A haplotypes were more in patients with T3-4 tumor size compared to T1-2 with respective value of OR = 3.61, CI = 1.10 - 11.8, P ≤ 0.01 and OR = 15.81, CI = 4.96 - 50.40, P < 0.01. With respect to metastasis, T-A haplotype was over-represented in M1 cases (OR: 3.98, CI = 1.25 - 12.6, P < 0.05). Further, C-A haplotype was elevated in < 6 months of DOD group with an OR of 0.24 (CI = 0.07 - 0.77, P < 0.01).
 Click to view | Table 6. Haplotype Analysis of CTLA-4 (rs11571317 and rs3087243) Gene Polymorphisms in Breast Cancer Patients and Controls and Haplotypes Association With Epidemiologic, Clinical Parameters in South Indian Breast Cancer Patients |
Hap map and LD analysis
The diversity of the allelic frequencies of CTLA4 rs11571317 and rs3087243 polymorphism in other populations was compared by Hap map data. The SNP rs11571317 wild-type “C” allele was observed to be predominant in all studied populations. Increased frequency of variant allele “T” was seen in the present study compared to Chinese, Asian (North India) and European populations as shown in Figure 3.
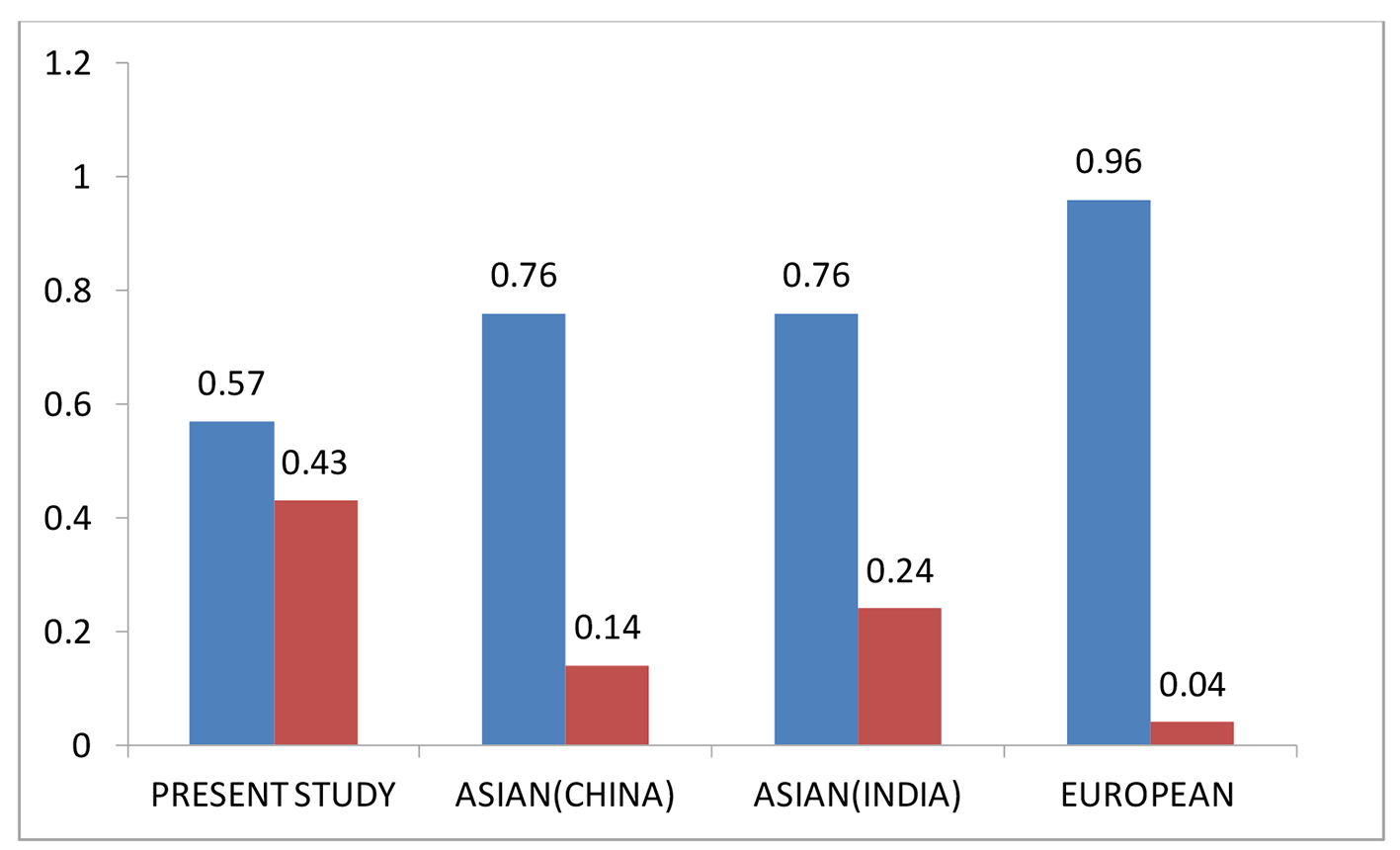 Click for large image | Figure 3. Comparative allele frequency of CTLA-4 SNP rs11571317 in various study populations. Source: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=11571317 |
The SNP rs3087243 with wild-type “G” allele was observed to be predominant in all the populations investigated. The frequency of the variant allele observed in the present study was similar to that of Asian population, whereas decreased frequency in African Americans, Sub Sahara populations and increased frequency in European population were noted, as shown in Figure 4.
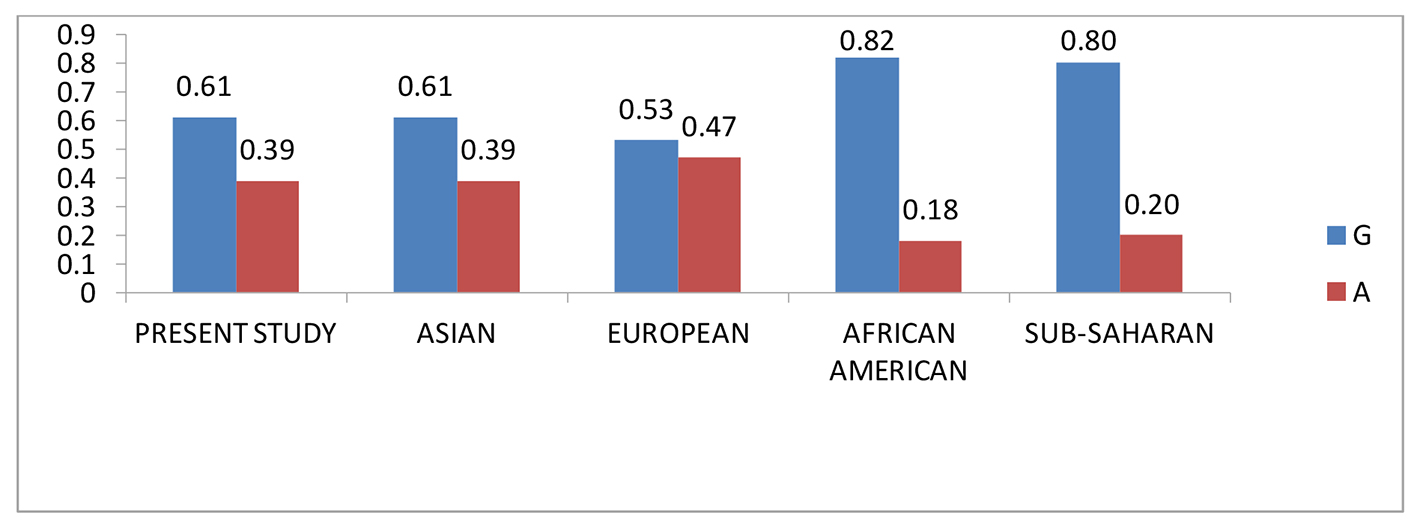 Click for large image | Figure 4. Comparative allele frequency of CTLA-4 SNP rs3087243 in various study populations. Source: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=3087243 |
Linkage disequilibrium plot of CTLA-4 gene for SNPs rs11571317 and rs3087243 in controls and patients exhibited LD value of 0.36, LOD of 0.48, and r-squared of 0.061, which suggested that these two SNPs act independently. Our LD results are in line with Maneesh et al (2014) in North Indian population.
| Discussion | ▴Top |
CTLA-4 is a key immune regulatory molecule that is involved in the immune homeostasis, associated with negative regulation of T-cell activation and tolerance. The genetic polymorphisms of this molecule may have their impact on transcriptional and translational aspects that might affect its function. Existence of correlation between CTLA-4 gene variants and alternative splicing has been demonstrated by Wang et al (2007). It was observed that the promoter and 3’UTR gene variants were known to influence the levels of expression of alternatively spliced (full-length isoform) flCTLA-4 and (soluble) sCTLA-4 isoform’s RNA [13, 23, 24]. The current study aimed to investigate the influence of a promoter polymorphism (rs11571317) and a 3’UTR SNP (rs3087243) in BC susceptibility and progression. We found CC genotype of rs11571317 to impart a three-fold increased risk towards BC, whereas the heterozygote CT was shown to be shielding. These observations of ours are supported by another study which also reported that CC genotype of this promoter polymorphism may increase BC risk and “T” allele to be protective in Han Chinese population. Further, our in-silico analysis revealed abolition of SP1 binding site in the CTLA-4 promoter by the mutant allele “T” suggesting low expression levels of CTLA-4. However, functional analysis should confirm these observations.
We noted neither risk nor protective genotypes for BC with respect to CTLA-4 SNP rs3087243. Similar results were reported by Wang et al (2007) in a case-control study where they found no variation in the frequency of the genotypes between patients and controls. When the analysis was carried out within the patients with respect to tumor growth, the wild-type GG genotype was significantly allied with restricted tumor growth, while GA and AA genotypes with increased tumor growth. This result of ours can be explained based on the report that the ratio of sCTLA-4 to flCTLA4 isoform mRNA splice forms in un-stimulated CD4 T cells is 50% lower in GG individuals compared with AA individuals postulating functional differences of CTLA-4 protein expression [13]. Thus, CT60 GG may confer a lesser CTLA4 function, resulting in greater T-cell activity, stronger immune response, and a higher probability of reduced tumor growth. On contrary to this, others showed lack of correlation between CT60 genotypes and serum sCTLA4 levels, even with higher serum sCTLA4 levels in CT60G alleles carriers of south-eastern United States [25] and in Graves’ disease patients of Poland [26].
In further analysis, T-G haplotype emerged as a protective haplotype in our population and C-A and T-A haplotypes were associated with increased tumor size. In relation to DOD, the C-A haplotype was more prevalent in patients with < 6 months of DOD compared to the group with > 6 months of DOD. Though both C-A and T-A haplotypes are involved in increased tumor growth, the effect of T-A haplotype was more profound (16-fold) than C-A (four-fold), indicating that the CT60 polymorphism may have a critical role compared to -658 C/T SNP in BC growth. Several investigators demonstrated that individuals with CT60 polymorphism had increased T regulatory cells and increased CTLA-4 levels; this information can serve as the basis for the augmented tumor growth as seen in our study [13, 27-29]. In addition, the over-representation of C-A haplotype in < 6 months DOD group is associated with restricted disease progression. Taken as a whole, alliance with increased tumor growth with C-A haplotype implies that sequential accelerated action of CTLA-4 SNPs is linked with initiation of disease to fully fledged tumor growth. In immune-related diseases, sCTLA-4 might compete with membrane bound CTLA-4 for CD80/CD86, and prevent down-regulation of T-cell response in the earlier stage of the disease, but it might inhibit T-cell activation by blocking the CD80/CD86-CD28 interaction in the advanced stage of the disease [30].
Identification of pivotal molecules in the stimulatory and inhibitory mechanisms of the immune system shows a potential role in the treatment of cancers. Our results suggest that the selected SNPs of CTLA-4 can serve as good bio and prognostic markers in our population. To the best of our knowledge, this is the first report on rs11571317 polymorphism with BC in Indian context. Large studies considering these SNPs and clinical and chemotherapeutic response in relation to soluble and membrane bound CTLA-4 will lead to better appliance of this molecule. The limitation of the study is lack of information on isoforms of CTLA-4 in serum and the tumor microenvironment.
Acknowledgments
We are grateful to the Osmania General Hospital, Railway Central Hospital, Soumya Multi-specialty Hospital and ESI Hospital, patients and controls for their cooperation in providing blood samples and clinical data. We thank the University Grant Commission (UGC), Department of Biotechnology-Interdisciplinary School of Life Sciences for Advanced Research and Education (DBT-ISLARE) and Centre for Advanced Studies-II (CAS-II) program for providing financial support.
Author Contributions
Parveen Jahan envisaged the manuscript, helped with manuscript drafting and gave critical inputs. Goske Maruthi and V. R. Vnish Ramachander together drafted and shaped the manuscript. Komaravalli Prasanna Latha provided valuable insights for drafting the manuscript and helped in statistics. P. Fazul Rahman encouraged us to manuscript preparation and S. Chandrasheker Rao helped out in samples collection.
Conflict of Interest
The authors declare that they have no conflict of interest.
| References | ▴Top |
- Jahan P, Ramachander VR, Maruthi G, Nalini S, Latha KP, Murthy TS. Foxp3 promoter polymorphism (rs3761548) in breast cancer progression: a study from India. Tumour Biol. 2014;35(4):3785-3791.
doi pubmed - Meraj Farbod, Seyed Mostafa Shiryazdi, Hamid Harazi, Tahereh Nazari, Mohammad Hasan Sheikhha. Association between the cytotoxic T-lymphocyte antigen-4 polymorphisms and breast cancer risk and prognosis. J Cancer Metastasis Treat. 2015;1:16-20.
doi - Vinish Ramachander VR, Maruthi G, Ranjith Reddy K, Siva Nagaprasad, Chandrasekhar Rao S, Parveen Jahan. Sway of FOXP3 gene variants in the genetic vulnerability of Asian Indian women towards breast cancer. International Journal of Advanced Scientific Research and Management. 2016;1(8).
- Gomez Flores-Ramos L, Escoto-De Dios A, Puebla-Perez AM, Figuera-Villanueva LE, Ramos-Silva A, Ramirez-Patino R, Delgado-Saucedo JI, et al. Association of the tumor necrosis factor-alpha -308G>A polymorphism with breast cancer in Mexican women. Genet Mol Res. 2013;12(4):5680-5693.
doi pubmed - Chen F, Zhou J, Xue Y, Yang S, Xiong M, Li Y, Liu Q. A single nucleotide polymorphism of the TNRC9 gene associated with breast cancer risk in Chinese Han women. Genet Mol Res. 2014;13(1):182-187.
doi pubmed - Yang Y, Wang W, Liu G, Yu Y, Liao M. Association of single nucleotide polymorphism rs3803662 with the risk of breast cancer. Sci Rep. 2016;6:29008.
doi pubmed - Welch DR, Steeg PS, Rinker-Schaeffer CW. Molecular biology of breast cancer metastasis. Genetic regulation of human breast carcinoma metastasis. Breast Cancer Res. 2000;2(6):408-416.
doi pubmed - Kohaar I, Tiwari P, Kumar R, Nasare V, Thakur N, Das BC, Bharadwaj M. Association of single nucleotide polymorphisms (SNPs) in TNF-LTA locus with breast cancer risk in Indian population. Breast Cancer Res Treat. 2009;114(2):347-355.
doi pubmed - Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31(1):33-36.
doi pubmed - Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet. 1992;2(2):128-131.
doi pubmed - Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet. 1995;56(1):265-271.
pubmed - Julian Kenrick Loh. Gene polymorphisms in the renin-angiotensin-aldosterone system and breast carcinogenesis: is there a connection? TSMJ. 2008;9:48-51.
- Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506-511.
doi pubmed - Fang W, Zhang Z, Zhang J, Cai Z, Zeng H, Chen M, Huang J. Association of the CTLA4 gene CT60/rs3087243 single-nucleotide polymorphisms with Graves' disease. Biomed Rep. 2015;3(5):691-696.
doi pubmed - Liu Z, Song Z, Sun J, Sun F, Li C, Sun J, Xu L. Association between CTLA-4 rs231775 polymorphism and hepatocellular carcinoma susceptibility. Int J Clin Exp Pathol. 2015;8(11):15118-15122.
pubmed - Wang L, Li D, Fu Z, Li H, Jiang W, Li D. Association of CTLA-4 gene polymorphisms with sporadic breast cancer in Chinese Han population. BMC Cancer. 2007;7:173.
doi pubmed - Misra MK, Kapoor R, Pandey SK, Sharma RK, Agrawal S. Association of CTLA-4 gene polymorphism with end-stage renal disease and renal allograft outcome. J Interferon Cytokine Res. 2014;34(3):148-161.
doi pubmed - Celmeli F, Turkkahraman D, Ozel D, Akcurin S, Yegin O. CTLA-4 (+49A/G) polymorphism and type-1 diabetes in Turkish children. J Clin Res Pediatr Endocrinol. 2013;5(1):40-43.
doi pubmed - Devaraju P, Gulati R, Singh BK, Mithun CB, Negi VS. The CTLA4 +49 A/G (rs231775) polymorphism influences susceptibility to SLE in South Indian Tamils. Tissue Antigens. 2014;83(6):418-421.
doi pubmed - Deeba F, Syed R, Quareen J, Waheed MA, Jamil K, Rao H. CTLA-4 A49G gene polymorphism is not associated with vitiligo in South Indian population. Indian J Dermatol. 2010;55(1):29-32.
doi pubmed - de Jong VM, Zaldumbide A, van der Slik AR, Laban S, Koeleman BP, Roep BO. Variation in the CTLA4 3'UTR has phenotypic consequences for autoreactive T cells and associates with genetic risk for type 1 diabetes. Genes Immun. 2016;17(1):75-78.
doi pubmed - Tippisetty S, Ishaq M, Komaravalli PL, Jahan P. Angiotensin converting enzyme (ACE) gene polymorphism in vitiligo: protective and predisposing effects of genotypes in disease susceptibility and progression. Eur J Dermatol. 2011;21(2):173-177.
pubmed - Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. 2006;24:65-97.
doi pubmed - Palacios R, Comas D, Elorza J, Villoslada P. Genomic regulation of CTLA4 and multiple sclerosis. J Neuroimmunol. 2008;203(1):108-115.
doi pubmed - Purohit S, Podolsky R, Collins C, Zheng W, Schatz D, Muir A, Hopkins D, et al. Lack of correlation between the levels of soluble cytotoxic T-lymphocyte associated antigen-4 (CTLA-4) and the CT-60 genotypes. J Autoimmune Dis. 2005;2:8.
doi pubmed - Daroszewski J, Pawlak E, Karabon L, Frydecka I, Jonkisz A, Slowik M, Bolanowski M. Soluble CTLA-4 receptor an immunological marker of Graves' disease and severity of ophthalmopathy is associated with CTLA-4 Jo31 and CT60 gene polymorphisms. Eur J Endocrinol. 2009;161(5):787-793.
doi pubmed - Atabani SF, Thio CL, Divanovic S, Trompette A, Belkaid Y, Thomas DL, Karp CL. Association of CTLA4 polymorphism with regulatory T cell frequency. Eur J Immunol. 2005;35(7):2157-2162.
doi pubmed - Karabon L, Kosmaczewska A, Bilinska M, Pawlak E, Ciszak L, Jedynak A, Jonkisz A, et al. The CTLA-4 gene polymorphisms are associated with CTLA-4 protein expression levels in multiple sclerosis patients and with susceptibility to disease. Immunology. 2009;128(1 Suppl):e787-796.
doi pubmed - Pincerati MR, Dalla-Costa R, Petzl-Erler ML. CTLA4CT60 gene polymorphism is not associated with differential susceptibility to pemphigus foliaceus. Genet Mol Biol. 2010;33(3):442-444.
doi pubmed - Saverino D, Brizzolara R, Simone R, Chiappori A, Milintenda-Floriani F, Pesce G, Bagnasco M. Soluble CTLA-4 in autoimmune thyroid diseases: relationship with clinical status and possible role in the immune response dysregulation. Clin Immunol. 2007;123(2):190-198.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


