| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website http://www.wjon.org |
Original Article
Volume 11, Number 1, February 2020, pages 1-8
Inclusion of an E7 DNA Amplification Test Improves the Robustness of Human Papillomavirus-Associated Oropharyngeal Squamous Cell Carcinoma Diagnosis
Pin-Yu Pereraa, d, Liyanage P. Pererab, Lyvouch Filkoskia, Wen Chena, Jack H. Lichya, Edina Paala, Jessica H. Maxwellc
aPathology and Laboratory Medicine, Veterans Affairs Medical Center, Washington, DC 20422, USA
bNational Cancer Institute, National Institutes of Health, Bethesda, MD, USA
cOtolaryngology, Georgetown University School of Medicine, Veterans Affairs Medical Center, Washington, DC, USA
dCorresponding Author: Pin-Yu Perera, Pathology and Laboratory Medicine, Veterans Affairs Medical Center, 50 Irving St., NW, Washington, DC 20422, USA
Manuscript submitted November 5, 2019, accepted December 10, 2019
Short title: PCR Improves HPV-OPSCC Diagnosis
doi: https://doi.org/10.14740/wjon1243
| Abstract | ▴Top |
Background: The rise in human papillomavirus (HPV) infection rates over the last few decades in the USA has contributed to a significant increase in the overall incidence of patients diagnosed with squamous cell carcinoma of the head and neck. These head and neck carcinomas develop in the oropharynx, with more than 90% of them caused by infection with high-risk HPV type 16. Patients diagnosed with HPV-induced oropharyngeal squamous cell carcinomas (OPSCCs) have a better prognosis and treatment response than those diagnosed with head and neck cancers caused by alcohol consumption and tobacco use. To identify patients with HPV-positive OPSCC, new guidelines recommend positive staining of oropharyngeal tissues for p16 INK4a (p16) by immunohistochemistry (IHC). Herein we discuss the testing algorithm that was adopted to address discrepant results between p16 IHC and a DNA in situ hybridization (ISH) test used routinely to diagnose HPV-positive OPSCC patients.
Methods: A DNA polymerase chain reaction (PCR) test that amplifies HPV16 and HPV18 E7 was developed to aid in the diagnosis of HPV-positive OPSCC in a subset of patients. Specimens from these patients stained positive for p16 by an IHC test, but negative for high-risk HPV by a commercial DNA ISH test. Moreover, these results did not match the histopathological characteristics of the specimens, nor the clinical presentations of the patients.
Results: Of 21 patients’ specimens that were tested for p16 by IHC, 11 specimens showed concordant results with the high-risk HPV 16/18 DNA ISH test. Whereas, in eight p16 IHC positive specimens, HPV viral DNA was not detected by HPV16/18 DNA ISH, and two specimens were not tested by DNA ISH. When these eight p16 IHC positive specimens with discrepant p16 IHC and DNA ISH results were further tested by DNA PCR, six specimens showed concordance with p16 IHC with positive results for HPV16 E7, while two specimens were negative for HPV16 E7 by DNA PCR. All tested specimens were negative for HPV18 E7 by DNA PCR. Thus, the addition of the HPV16 and HPV18 E7 DNA PCR test identified a significant number of false negative test results by the HPV16/18 DNA ISH test and likely several false positive results by p16 IHC.
Conclusions: Inclusion of an HPV16 E7 DNA PCR test improved the robustness of HPV-associated OPSCC diagnosis in patients with discrepant results from p16 IHC staining and a DNA ISH test, and identified patients for proper management with less misclassification.
Keywords: Oropharyngeal squamous cell carcinoma; Human papillomavirus; Polymerase chain reaction; p16 immunohistochemistry; DNA in situ hybridization; HPV E7 DNA amplification
| Introduction | ▴Top |
Human papillomavirus (HPV) infection is the most common sexually transmitted infection in the USA [1]. Coincident with the HPV epidemic there is a steady rise in a subset of head and neck cancers arising in the oropharynx comprised of lingual and palatine tonsils, the base of the tongue, lateral and posterior pharyngeal walls, and soft palate [2, 3]. In contrast, there is a marked decline in head and neck cancers arising in the oral cavity, larynx and the hypopharynx caused by alcohol and carcinogens in tobacco [4, 5]. Majority of individuals infected with HPV are asymptomatic and the infections are cleared naturally by the immune system, although men are reportedly less likely to clear an infection than women [6-8]. The average latency period to develop HPV-positive oropharyngeal squamous cell carcinomas (OPSCCs) is approximately 10 to 30 years [8]. HPV-positive OPSCCs are more likely to develop in younger Caucasian men with a light or no history of tobacco use compared to their HPV-negative counterparts [9, 10]. In addition, more than 80% of HPV-positive OPSCC patients present with advanced stage disease [11]. It is now well established that up to 70% of OPSCC in the USA is caused by sexually transmitted HPV virus due to changes in sexual norms and behavior patterns [12]. While more than 200 HPV types are known, only 13 to 15 high-risk HPV types are oncogenic, with HPV16 causing more than 90% of these OPSCCs, and to a lesser extent several other high-risk types namely 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 73 [13, 14].
HPV is a small non-enveloped circular double stranded DNA virus that encodes three oncoproteins E5, E6, and E7 [15]. Following an HPV infection, the viral oncoprotein E6 causes degradation of the p53 tumor suppressor protein that leads to genomic instability [16, 17]. The viral oncoprotein E7 binds the RB tumor suppressor protein and prevents negative feedback control of a cell cycle regulatory protein p16, thereby resulting in p16 overexpression [18, 19]. The combined effects of genomic instability and uncontrolled cell proliferation lead to cellular transformation and malignancy [16, 18]. Because p16 overexpression is associated with transcriptionally active HPV oncoprotein E7 and functional inactivation of the RB protein, overexpressed p16 is often used as a surrogate marker of active HPV infection [19].
While it is well recognized that HPV-positive patients need to be identified for proper management of these patients, no guidelines were available that detailed the methods to be used for HPV detection in clinical specimens until recently [20, 21]. In 2018, new guidelines published for HPV testing in head and neck carcinomas by the College of American Pathologists (CAP) strongly recommend high-risk HPV testing on all patients with newly diagnosed OPSCC, to include all histologic subtypes [22]. Moreover, these guidelines recommend high-risk HPV testing to be performed on oropharyngeal tissue specimens by IHC for the surrogate marker p16 and for HPV type-specific testing to be done at the discretion of the pathologist [22].
This study describes a polymerase chain reaction (PCR)-based test that was developed to aid in the diagnosis of HPV-positive OPSCC caused by high-risk HPV types 16 and 18 responsible for the majority of cases in the USA. The HPV16 E7 and HPV18 E7 PCR test was used as an alternative test method to increase diagnostic accuracy when discrepant results were obtained by p16 immunostaining and HPV DNA in situ hybridization (ISH).
| Materials and Methods | ▴Top |
Detection of p16 proteins by immunohistochemistry (IHC)
Four microns sections of formalin fixed paraffin embedded (FFPE) tissues on positively charged slides were deparaffinized with xylene and rehydrated with graded alcohols. Following antigen retrieval with Tris, pH 8.8 - 9.4 (PT Link, Dako, Agilent, Santa Clara, CA), and endogenous peroxide block with hydrogen peroxide, tissue sections were incubated with mouse monoclonal anti-p16INK4a antibody clone E6H4 (Roche Diagnostics, Indianapolis, IN). A tissue section incubated with normal mouse immunoglobulin G (IgG) antibody and a previously identified strongly p16 immune reactive patient tissue section were included in each run as negative and positive controls respectively. Immune reactive p16 positive cells were detected with Envision dual link system polymer containing goat anti-mouse secondary antibody conjugated to horse radish peroxidase (Dako, Agilent, Santa Clara, CA) and substrate 3, 3’-diaminobenzidine as chromogen. The nuclei were counterstained with hematoxylin, tissue slides were then dehydrated with alcohol, cleared in xylene and mounted. This procedure was performed on an automated instrument, Dako Autostainer Link 48. Immune reactivity for p16 was evaluated by staff pathologists. Tissues were scored as positive, equivocal or negative for p16.
Detection of HPV DNA by ISH
This test was performed by a commercial reference laboratory on FFPE tissue specimens. Briefly, probe mixes for HPV 6 and 11 (ENZ-3285), HPV 16 and 18 (ENZ-3286) were purchased (Enzo Life Sciences, Inc., Farmingdale, NY). The 2,4-dinitrophenyl (DNP) labeled probes hybridized to specific HPV target sequences in the tissue sections were detected with an anti-DNP antibody; followed by an indirect biotin-streptavidin-alkaline phosphatase system (Ventana ISH iViewBlue Plus Detection System, catalog number 760-097, Roche, Indianapolis, IN) with nitro-blue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as substrate and chromogen respectively. Tissue sections were counter stained with neutral red. A blue colored precipitate detected by light microscopy at sites where HPV probes hybridized was interpreted as positive for HPV. This procedure was performed on the Ventana BenchMark fully automated slide stainer (Roche, Indianapolis, IN). Reports were received as HPV high risk or HPV low risk detected or not detected.
DNA extraction from FFPE tissue specimens
DNA was extracted from FFPE tissues using the QiaAamp DNA FFPE tissue kit (Qiagen, Valencia, CA). Briefly, sections (4 - 5 µm) of FFPE tissues were cut from each tissue block and deparaffinized with CitriSolv (Fisher Scientific, Pittsburgh, PA). DNA was then extracted manually according to the manufacturer’s instructions using proteinase K digestion overnight. Following extraction, DNA concentration and quality were assessed using a Nanodrop spectrophotometer.
Detection of HPV16 E7 DNA, HPV18 E7 DNA, and human beta actin by PCR
To detect HPV viral DNA, primers for HPV16 E7 and HPV18 E7 were multiplexed to yield an amplicon of 220 base pairs for HPV16 E7 and an amplicon of 252 base pairs for HPV18 E7, by PCR target template amplification. The sequences of the HPV primers used are as follows: HPV16 E7 forward primer: 5' ATG GAG ATA CAC CTA CAT TGC ATG 3'; HPV16 E7 reverse primer: 5’ TCT ACG TGT GTG CTT TGT ACG C 3'; HPV18 E7 forward primer: 5' GAG CCC CAA AAT GAA ATT CCG G 3'; HPV18 E7 reverse primer: 5' CGG ACA CAC AAA GGA CAG GGT 3'. PCR was performed in a 50 µL reaction volume, with 25 µL of platinum multiplex PCR master mix (Life Technologies, Thermo Fisher Scientific, Wilmington, DE), 10 µL of HPV16 and HPV18 E7 primer mix (each primer at 0.5 µM concentration) and 15 µL of extracted DNA (25 ng to 500 ng per reaction) and water. The PCR reactions were initiated by activation of platinum DNA polymerase at 95 °C for 2 min, followed by 40 cycles of denature at 95 °C for 30 s, anneal at 60 °C for 30 s, and extend at 72 °C for 30 s, with a final extension at 72 °C for 10 min and hold at 4 °C. Water was used as a no template control in each run. Forty cycles were performed to produce a sufficient yield of PCR products to enable the detection of fewer than 10 copies of HPV E7 DNA as recommended by ThermoFisher (PCR cycle number determination, https://www.thermofisher.com/us). A previously confirmed HPV16 E7 positive FFPE DNA patient sample and pCRII-TOPO vector cloned with HPV18 E7 DNA were used as positive controls. To detect human beta actin, PCR was performed in a 50 µL reaction volume, with a PCR master mix that contained 1.25 units of AmpliTaq Gold DNA polymerase (Life Technologies, Thermo Fisher Scientific, Wilmington, DE), 1 × PCR buffer, 2.5 mM MgCl2, 200 µM of each deoxynucleotide triphosphate, and 0.2 µM of each beta actin primer and genomic DNA (2 ng to 250 ng). The PCR reactions were initiated by activation of Amplitaq gold DNA polymerase at 95 °C for 5 min, followed by 40 cycles of denature at 95 °C for 30 s, anneal at 57 °C for 30 s, and extend at 72 °C for 30 s, with a final extension at 72 °C for 10 min and hold at 4 °C. The following beta actin primer set generated an amplicon of 227 base pairs in size. Human beta actin forward primer: 5' TGC TAT CCC TGT ACG CCT CT 3'; human beta actin reverse primer: 5' CTC CTT AAT GTC ACG CAC GA 3'. To size the products, the PCR amplified products and a 100 base pairs ladder (Bio Rad Laboratories, Hercules, CA) were then electrophoresed on a 2% agarose Tris-acetate-EDTA (ethylenediamine tetraacetic acid) gel with ethidium bromide and the amplified products were viewed for the correct product size on a ultraviolet transilluminator. Images of the gels were acquired using the GE ImageQuant LAS4010 instrument.
DNA sequencing of HPV16 E7, HPV18 E7 and human beta actin genes
To sequence HPV16 and HPV18 E7 viral DNA and human beta actin DNA, PCR amplified products were purified using DNA clean and concentrator-5 columns (Zymo Research, Irvine, CA). Cycle sequencing was then performed on a GeneAmp 9700 PCR instrument using the Big Dye Terminator Ready Reaction cycle sequencing kit vs1.1 (Applied Biosystems, Foster City, CA). Each reaction contained purified PCR products as template DNA with the same gene specific forward or reverse primer (3.2 pmol per reaction) used for PCR, 2 µL of ready reaction premix and 1 µL Big Dye sequencing buffer in a total reaction volume of 10 µL per reaction. Cycle sequencing was then performed with an initial denaturation at 96 °C for 1 min, followed by 40 cycles of 96 °C for 10 s, 50 °C for 10 s and 60 °C for 4 min and hold at 4 °C. Following cycle sequencing the primer extension products were purified by ethanol/EDTA precipitation, denatured and the single stranded DNA fragments separated by capillary electrophoresis on an ABI 3130xl genetic analyzer. The sequenced nucleotides were then viewed with the ABI sequencing analysis software version 5.2. Basic Local Alignment Search Tool, (BLAST, NCBI, NIH) DNA alignment searches were then performed to confirm the identity of HPV16 and HPV18 E7 nucleotide sequences in the tumor specimens or positive controls [23]. Similar BLAST DNA alignment searches were performed with sequenced nucleotides generated for human beta actin to confirm the presence of beta actin in specimens.
| Results | ▴Top |
Clinical samples and test algorithm
Male patients with tumors in the head and neck with a clinical presentation suggestive of HPV infection were tested for active HPV infection as follows. Following initial histopathological review of hematoxylin and eosin (H&E)-stained FFPE tumor tissue specimens for squamous cell carcinoma morphology by a staff pathologist, FFPE tissue sections were tested for active HPV infection by IHC staining for the surrogate HPV marker p16. A specimen from a tonsil with p16 focal positivity that did not meet CAP guidelines for positivity and was scored p16 negative (Fig. 1) and a specimen from the base of the tongue that met CAP guidelines for positivity and was scored as p16 positive by the pathologists is shown in Figure 2. Specimens that tested positive for p16 were then confirmed for HPV infection. FFPE tissue blocks were sent out to a commercial reference laboratory to detect the presence of high risk (HPV16 and HPV18) or low risk (HPV6 and HPV11) HPV types by a proprietary DNA ISH test. Because of discordant p16 IHC and DNA ISH test results, specimens were also further tested by an inhouse newly developed PCR test to detect HPV16 E7 and HPV18 E7 DNA. A representative image of an agarose gel electrophoresed with PCR amplified HPV16 E7 in a patient sample, with HPV16 and HPV18 positive controls and negative FFPE DNA extraction and no template controls is shown in Figure 3. During assay validation the presence of HPV16 E7 DNA in the HPV PCR amplified positive samples was confirmed by direct sequencing for HPV16 E7 in forward and reverse directions and the generated nucleotide sequences were found to match HPV16 E7 with 100% identity by BLAST (NCBI, NIH) searches.
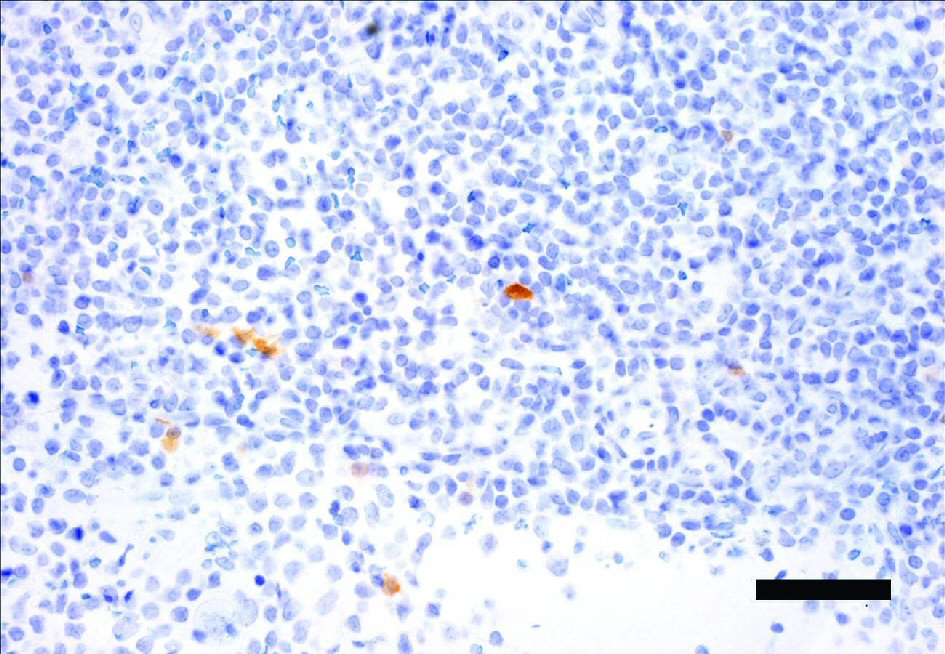 Click for large image | Figure 1. Tonsil specimen immunostained for p16. FFPE section from a tonsil scored negative for p16, with few focally positive stained cells. Magnification bar = 50 µm. FFPE: formalin fixed paraffin embedded. |
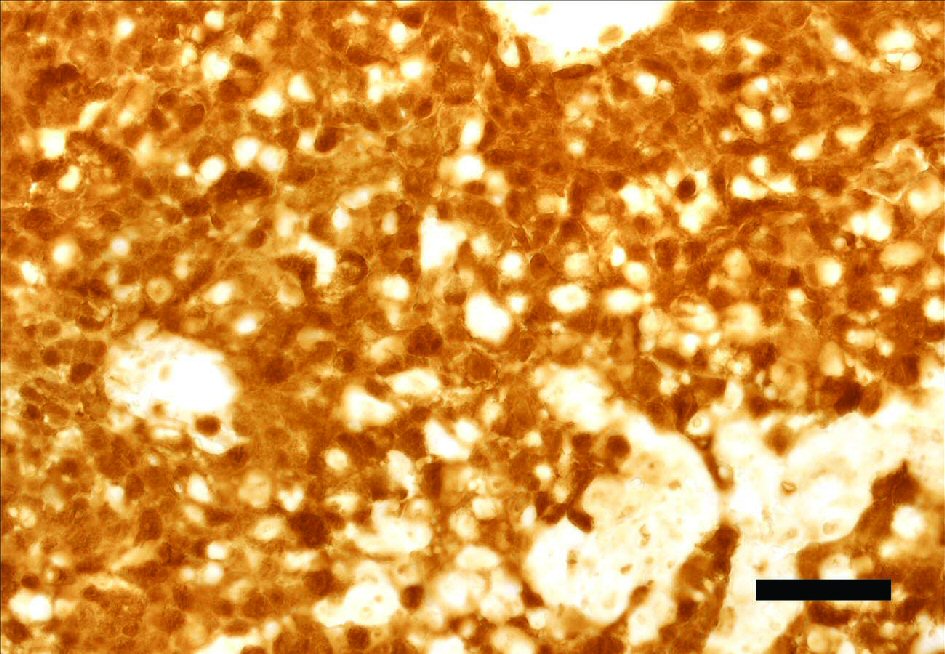 Click for large image | Figure 2. Base of the tongue specimen immunostained for p16. FFPE section from the base of the tongue scored positive for p16, with greater than 70% of nuclei and cytoplasm of tumor cells stained positive. Magnification bar = 50 µm. FFPE: formalin fixed paraffin embedded. |
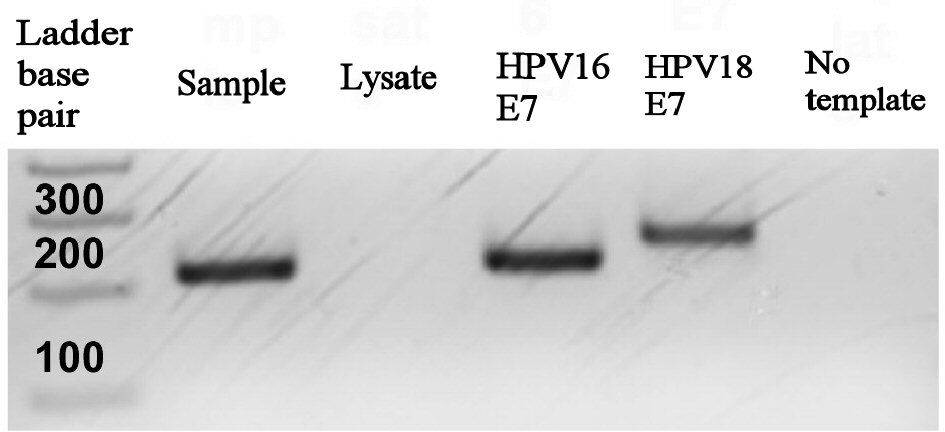 Click for large image | Figure 3. Gel electrophoresis of HPV16 E7 and HPV18 E7 PCR amplicons. Indicated samples were electrophoresed with a 100 base pair amplicon sizing ladder on a 2% agarose gel containing ethidium bromide. HPV: human papillomavirus; PCR: polymerase chain reaction. |
Test results
Table 1 shows the patient and tumor specimen characteristics. A total of 21 patient specimens were tested to date by the above outlined diagnostic algorithm. Of the specimens tested, 18 tested positive for p16 by immunostaining, while one specimen from a tonsil, the floor of the mouth and hypopharynx tested negative for p16 (Table 2). When the 18 p16 positive specimens were tested for the presence of active HPV16/18 by DNA ISH testing, nine specimens tested positive and eight specimens tested negative for high-risk HPV types, with one lymph node specimen and a specimen from the floor of the mouth not tested, while all specimens tested negative for low-risk HPV6 and HPV11.
 Click to view | Table 1. Patient and Tumor Specimen Characteristics |
 Click to view | Table 2. Results of p16 Immunostaining, High-Risk (HPV16/18) DNA ISH, and DNA PCR for HPV16 E7 for Each Patient Specimen Tested |
Of the 21 specimens that were tested for HPV16 E7 and HPV18 E7 DNA by multiplex PCR (Table 2), 17 specimens tested positive for HPV16 E7 DNA and four specimens tested negative for HPV16 E7 DNA, while all specimens tested negative for HPV18 E7 DNA except the positive control plasmid with cloned HPV18. All four HPV negative specimens amplified human beta actin indicating the absence of PCR inhibitors in the extracted DNA from FFPE tissue specimens and confirmed true absence of HPV16 and HPV18 DNA in these tumor tissues.
Based on our testing algorithm, 11 specimens showed concordance with high-risk HPV 16/18 DNA ISH and HPV16 E7 DNA PCR assay results, with nine specimens testing positive for HPV and two specimens testing negative for HPV. Six specimens had discordant results, testing negative for high-risk HPV16/18 by DNA ISH, while testing positive for HPV16 by DNA PCR and p16 IHC. The HPV16 E7 positivity was confirmed by sequencing all six discordant samples. Three specimens had discordant results by p16 IHC and HPV16 by DNA PCR.
This study was reviewed by the local institutional review board and was exempted from the requirement for informed consent.
| Discussion | ▴Top |
HPV-positive OPSCC patients typically respond well to concurrent platinum-based chemotherapy and radiation treatment [24]. However, patients often suffer from treatment-related side effects, including acute toxicity with chemotherapy and late toxicities such as xerostomia and dysphagia in response to high dose radiation [25, 26]. Clinical trials have identified a subpopulation of low-risk HPV positive-OPSCC patients including those with low volume tumors, minimal smoking history and less bulky cervical lymphadenopathy [20, 26]. This subgroup of patients may benefit from reduced dose radiation therapy that would reduce late toxicities and improve overall nutritional status [20, 27]. Thus, the importance of identifying HPV-positive OPSCC patients who have a clinically and molecularly distinct disease with better overall survival than HPV-negative patients is apparent from the new guidelines released by CAP for diagnosing HPV-positive OPSCC [22].
The eighth edition of the American Joint Committee on Cancer (AJCC) cancer staging manual, with TNM classification from the AJCC and the Union for International Cancer Control (UICC) has incorporated HPV into the staging system for oropharyngeal cancers [28, 29]. The staging of patients with OPSCC is now highly dependent on the HPV status, and those with HPV-positive OPSCC are often down-staged compared to their HPV-negative OPSCC counterparts [28, 29]. Furthermore, the pathologic staging differs from clinical staging by including the metastatic node count after a neck dissection [28]. Interestingly, the eighth edition AJCC staging system is based on a study conducted by O’Sullivan et al and the International Collaboration on Oropharyngeal Cancer Network for Staging (ICON-S), which determined HPV positivity by HPV-DNA ISH or by p16 IHC [30]. That the new guidelines released by CAP for diagnosing HPV positive OPSCC strongly recommend testing oropharyngeal tissue specimens for high-risk HPV by testing for the surrogate marker p16 by IHC is not surprising given the wide availability of IHC testing in most pathology laboratories, in addition to the cost effectiveness and ease of set up of IHC tests when compared to molecular-based technologies.
In the absence of published guidelines for HPV testing of head and neck cancers until recently, routine testing for HPV-positive head and neck cancers at our medical center included testing for the surrogate marker p16 by immunostaining. These results were then confirmed by DNA ISH for high-risk HPV16/18 in a commercial reference laboratory. However, this testing resulted in patients with clinical presentations strongly suggestive of HPV-related OPSCC who tested positive for p16 but tested negative for high-risk HPV16/18 by DNA ISH. While DNA ISH maintains the tissue specimen morphology that is essential for histopathological interpretation, the sensitivity of the proprietary ISH test even with signal amplification appeared to be low. Furthermore, interpretation is also reported to be challenging in ISH tests when episomal DNA masks few integrated forms of HPV present in the same cell [31]. To rule out false negative HPV16 and HPV18 cases by ISH, a new PCR test was established to detect HPV16 and HPV18 E7 DNA in FFPE tissues. HPV E7 was selected as the PCR target because of its role as the most potent HPV oncogene that is integrated, whereas E5 is lost during genomic integration [32]. Although several Food and Drug Administration (FDA)-approved commercial tests are available for detecting high-risk HPV in cervical specimens, currently no FDA-approved tests are available for detecting high-risk HPV in OPSCC [33]. While the detection of HPV E7 RNA would be more indicative of a persistent infection associated with a carcinoma rather than a transient infection, a DNA test was developed instead of an RNA test due to the unreliability of extracting high quality RNA from FFPE tissues.
In the cases tested, two specimens that tested positive for p16 by IHC were negative for active HPV 16/18 when tested by HPV DNA ISH and by HPV DNA PCR. Both specimens were from the base of tongue. Since the presence of PCR inhibitors was ruled out, these results suggest that patients were either infected with one of the rarer high-risk HPV types that was not tested for, or that p16 IHC was false positive due to the subjective nature of scoring prior to the published guidelines. The current guidelines for p16 IHC positive scoring require tumors scored as positive for p16 to stain positive in greater than 70% of the nuclei and the cytoplasm of carcinoma cells in the tissue section [22]. Another review of discrepant tissue sections for p16 IHC positivity based on the new guidelines confirmed the previous subjective nature of positive scoring when two samples with focal staining failed to meet the criteria for positivity set by the new guidelines. A false positive rate of up to 7.3% has been previously reported for p16 IHC staining [34]. In addition, p16 expression and p16-positive IHC staining are not specific for HPV infections, since mutations in the RB gene would lead to a similar increase in p16 expression [35]. Interestingly, in a large Dutch cohort of oropharyngeal cancer involving 1,204 patients from two medical centers, where tumor samples underwent p16 IHC staining and positive specimens were additionally tested for high-risk HPV-DNA by PCR, a subset of patients were identified as p16-positive and HPV DNA negative [35, 36]. Thus, if based solely on p16 status 12% of these Dutch patients would be incorrectly classified as having HPV-positive cancers [36, 37]. Based on the high sensitivity of our PCR test able to detect fewer than 10 copies of HPV viral DNA, six specimens from tumors located in the oropharynx that were p16 positive but negative for HPV by DNA ISH were identified as HPV16 positive by PCR. These results have direct clinical implications for these HPV-positive patients since they are down-staged with better prognosis, require proper counseling and follow-up surveillance imaging based on their HPV-positive status. While the treatment regimen does not change for HPV-positive patients who have a good prognosis with established treatment, they can potentially enroll in treatment de-escalation or de-intensification clinical trials that consider reduction in radiation dose following induction chemotherapy, radiotherapy alone, minimally invasive surgical techniques, or substituting platinum-based chemotherapy [20, 21, 27, 38]. According to the new CAP guidelines, a tumor located on the floor of the mouth that tested negative for p16 but was positive for HPV16 by DNA PCR, did not require HPV testing due to the lack of evidence to support a better prognosis for an HPV-positive primary tumor originating at a site other than the oropharynx [22]. Similarly, one cervical lymph node metastasis with an HPV positive primary located in the oropharynx did not require repeat HPV testing. No HPV testing was also required for a tumor located in the hypopharynx. Furthermore, no testing is required for non-oncogenic HPV6 and HPV11.
While the small sample size is a limitation of the study, a larger retrospective study found that p16 IHC alone performed less well than when the HPV status of the patients were determined by two methods, either HPV-DNA and p16 IHC or HPV mRNA [39]. This larger study was performed on HPV-positive oropharyngeal patients from four hospitals in Catalonia to identify the best HPV detection method that would correctly determine the overall survival in patients according to the eighth edition AJCC TNM classifier [39].
In summary, we have developed a rapid HPV16 E7 DNA PCR test that can be performed in 2 to 3 days with equipment that is available in most molecular laboratories and is easy to adopt inhouse. The test is sensitive, simpler to evaluate with less subjective interpretation than seen in HPV DNA ISH and p16 IHC. This test can be used to aid in the diagnosis of the most common HPV16 type responsible for OPSCC, in instances where results are ambiguous and need further clarification for diagnostic accuracy.
Acknowledgments
We thank Jean Przybocki for DNA extractions and Dr. Robert Redman for his expert help in preparing the p16 IHC figures.
Financial Disclosure
This publication is based upon work supported by the Department of Veterans Affairs and in part from awarded grant support (PYP) by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001409. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Veterans Affairs or the National Institutes of Health.
Conflict of Interest
The authors have declared no conflict of interest.
Informed Consent
This study was reviewed by the local institutional review board and was exempted from the requirement for informed consent.
Author Contributions
PYP developed the PCR and sequencing assays, generated data, and wrote the manuscript. LPP provided cloned HPV plasmids, contributed to assay development and edited the manuscript. LF performed p16 IHC. JL developed the concept for the study. WC and EP evaluated the p16 IHC slides. JM wrote the final revision. All the authors read and approved the final version of this manuscript.
| References | ▴Top |
- Satterwhite CL, Torrone E, Meites E, Dunne EF, Mahajan R, Ocfemia MC, Su J, et al. Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sex Transm Dis. 2013;40(3):187-193.
doi pubmed - Corey AS, Hudgins PA. Radiographic imaging of human papillomavirus related carcinomas of the oropharynx. Head Neck Pathol. 2012;6(Suppl 1):S25-40.
doi pubmed - Haeggblom L, Ramqvist T, Tommasino M, Dalianis T, Nasman A. Time to change perspectives on HPV in oropharyngeal cancer. A systematic review of HPV prevalence per oropharyngeal sub-site the last 3 years. Papillomavirus Res. 2017;4:1-11.
doi pubmed - Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer. 2005;114(5):806-816.
doi pubmed - Denson L, Janitz AE, Brame LS, Campbell JE. Oral cavity and oropharyngeal cancer: changing trends in incidence in the United States and Oklahoma. J Okla State Med Assoc. 2016;109(7-8):339-345.
- Kreimer AR, Pierce Campbell CM, Lin HY, Fulp W, Papenfuss MR, Abrahamsen M, Hildesheim A, et al. Incidence and clearance of oral human papillomavirus infection in men: the HIM cohort study. Lancet. 2013;382(9895):877-887.
doi - Beachler DC, Sugar EA, Margolick JB, Weber KM, Strickler HD, Wiley DJ, Cranston RD, et al. Risk factors for acquisition and clearance of oral human papillomavirus infection among HIV-infected and HIV-uninfected adults. Am J Epidemiol. 2015;181(1):40-53.
doi pubmed - Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J Clin Oncol. 2015;33(29):3235-3242.
doi pubmed - Deschler DG, Richmon JD, Khariwala SS, Ferris RL, Wang MB. The "new" head and neck cancer patient-young, nonsmoker, nondrinker, and HPV positive: evaluation. Otolaryngol Head Neck Surg. 2014;151(3):375-380.
doi pubmed - Cole L, Polfus L, Peters ES. Examining the incidence of human papillomavirus-associated head and neck cancers by race and ethnicity in the U.S., 1995-2005. PLoS One. 2012;7(3):e32657.
doi pubmed - Dahlstrom KR, Anderson KS, Sturgis EM. Human papillomavirus-associated oropharyngeal cancer: not just white men anymore. JAMA Oncol. 2017;3(2):161-162.
doi pubmed - Rettig E, Kiess AP, Fakhry C. The role of sexual behavior in head and neck cancer: implications for prevention and therapy. Expert Rev Anticancer Ther. 2015;15(1):35-49.
doi pubmed - Burd EM. Human papillomavirus laboratory testing: the changing paradigm. Clin Microbiol Rev. 2016;29(2):291-319.
doi pubmed - Marur S, D'Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11(8):781-789.
doi - Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res. 2009;15(22):6758-6762.
doi pubmed - Slebos RJ, Lee MH, Plunkett BS, Kessis TD, Williams BO, Jacks T, Hedrick L, et al. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci U S A. 1994;91(12):5320-5324.
doi pubmed - Munger K, Scheffner M, Huibregtse JM, Howley PM. Interactions of HPV E6 and E7 oncoproteins with tumour suppressor gene products. Cancer Surv. 1992;12:197-217.
- Khleif SN, DeGregori J, Yee CL, Otterson GA, Kaye FJ, Nevins JR, Howley PM. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci U S A. 1996;93(9):4350-4354.
doi pubmed - Gronhoj Larsen C, Gyldenlove M, Jensen DH, Therkildsen MH, Kiss K, Norrild B, Konge L, et al. Correlation between human papillomavirus and p16 overexpression in oropharyngeal tumours: a systematic review. Br J Cancer. 2014;110(6):1587-1594.
doi pubmed - Chen AM, Felix C, Wang PC, Hsu S, Basehart V, Garst J, Beron P, et al. Reduced-dose radiotherapy for human papillomavirus-associated squamous-cell carcinoma of the oropharynx: a single-arm, phase 2 study. Lancet Oncol. 2017;18(6):803-811.
doi - Hegde JV, Shaverdian N, Felix C, Wang PC, Veruttipong D, Hsu S, Riess JW, et al. Functional outcomes after de-escalated chemoradiation therapy for human papillomavirus-positive oropharyngeal cancer: secondary analysis of a phase 2 trial. Int J Radiat Oncol Biol Phys. 2018;100(3):647-651.
doi pubmed - Lewis JS, Jr., Beadle B, Bishop JA, Chernock RD, Colasacco C, Lacchetti C, Moncur JT, et al. Human papillomavirus testing in head and neck carcinomas: guideline from the college of American pathologists. Arch Pathol Lab Med. 2018;142(5):559-597.
doi pubmed - Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403-410.
doi - Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261-269.
doi pubmed - Chera BS, Fried D, Price A, Amdur RJ, Mendenhall W, Lu C, Das S, et al. Dosimetric predictors of patient-reported xerostomia and dysphagia with deintensified chemoradiation therapy for HPV-associated oropharyngeal squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2017;98(5):1022-1027.
doi pubmed - Caudell JJ, Schaner PE, Meredith RF, Locher JL, Nabell LM, Carroll WR, Magnuson JS, et al. Factors associated with long-term dysphagia after definitive radiotherapy for locally advanced head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2009;73(2):410-415.
doi pubmed - Marur S, Li S, Cmelak AJ, Gillison ML, Zhao WJ, Ferris RL, Westra WH, et al. E1308: Phase II Trial of Induction Chemotherapy Followed by Reduced-Dose Radiation and Weekly Cetuximab in Patients With HPV-Associated Resectable Squamous Cell Carcinoma of the Oropharynx- ECOG-ACRIN Cancer Research Group. J Clin Oncol. 2017;35(5):490-497.
doi pubmed - Lydiatt WM, Patel SG, O'Sullivan B, Brandwein MS, Ridge JA, Migliacci JC, Loomis AM, et al. Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(2):122-137.
doi pubmed - Cramer JD, Hicks KE, Rademaker AW, Patel UA, Samant S. Validation of the eighth edition American Joint Committee on Cancer staging system for human papillomavirus-associated oropharyngeal cancer. Head Neck. 2018;40(3):457-466.
doi pubmed - O'Sullivan B, Huang SH, Su J, Garden AS, Sturgis EM, Dahlstrom K, Lee N, et al. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol. 2016;17(4):440-451.
doi - Guo M, Gong Y, Deavers M, Silva EG, Jan YJ, Cogdell DE, Luthra R, et al. Evaluation of a commercialized in situ hybridization assay for detecting human papillomavirus DNA in tissue specimens from patients with cervical intraepithelial neoplasia and cervical carcinoma. J Clin Microbiol. 2008;46(1):274-280.
doi pubmed - Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun. 2013;4:2513.
doi pubmed - Leal SM, Jr., Gulley ML. Current and emerging molecular tests for human papillomavirus-related neoplasia in the genomic era. J Mol Diagn. 2017;19(3):366-377.
doi pubmed - Seiwert T. Accurate HPV testing: a requirement for precision medicine for head and neck cancer. Ann Oncol. 2013;24(11):2711-2713.
doi pubmed - Hayes DN, Van Waes C, Seiwert TY. Genetic Landscape of Human Papillomavirus-Associated Head and Neck Cancer and Comparison to Tobacco-Related Tumors. J Clin Oncol. 2015;33(29):3227-3234.
doi pubmed - Orosco RK, Califano JA. HPV status, like politics, is local-evaluating p16 staining and a new staging system in a Dutch cohort of oropharynx cancer. Ann Oncol. 2018;29(5):1089-1090.
doi pubmed - Nauta IH, Rietbergen MM, van Bokhoven A, Bloemena E, Lissenberg-Witte BI, Heideman DAM, Baatenburg de Jong RJ, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol. 2018;29(5):1273-1279.
doi pubmed - Kelly JR, Husain ZA, Burtness B. Treatment de-intensification strategies for head and neck cancer. Eur J Cancer. 2016;68:125-133.
doi pubmed - Taberna M, Mena M, Tous S, Pavon MA, Oliva M, Leon X, Garcia J, et al. HPV-relatedness definitions for classifying HPV-related oropharyngeal cancer patient do impact on TNM classification and patients' survival. PLoS One. 2018;13(4):e0194107.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


