| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 13, Number 4, August 2022, pages 205-215
N6-Methyladenosine RNA Modification Landscape in the Occurrence and Recurrence of Nasopharyngeal Carcinoma
Yu Min Wanga, b, c, e, Zhou Ying Penga, b, c, e, Lu Yuan Zhangd, Ya Xuan Wanga, b, Ruo Hao Fana, b, c, Hua Zhanga, b, c, Wei Hong Jianga, b, c, f
aDepartment of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, China
bNational Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Changsha, China
cAnatomy Laboratory of Division of Nose and Cranial Base, Clinical Anatomy Center of Xiangya Hospital, Central South University, Changsha, China
dDepartment of Neurosurgery, First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China
eThese authors contributed equally.
fCorresponding Author: Wei Hong Jiang, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, China
Manuscript submitted May 19, 2022, accepted June 24, 2022, published online August 23, 2022
Short title: m6A in Occurrence and Recurrence of NPC
doi: https://doi.org/10.14740/wjon1491
| Abstract | ▴Top |
Background: Nasopharyngeal carcinoma (NPC) is a type of squamous head and neck cancer with variable geographic distributions, with the highest incidence in Southeast Asia. Its primary treatment is radiotherapy due to its high radio sensitivity. However, the N6-methyladenosine (m6A) landscape in NPC, including recurrent NPC, has not been reported.
Methods: In this study, m6A RNA immunoprecipitation (RIP) sequencing and microarray sequencing were performed on 12 tissue samples tissues of patients with primary and recurrent NPC. The expression profiles of m6A-related and non-coding RNAs were constructed and explored. Then, function experiments were performed to evaluate the effects of methyltransferase (METTL)3, METTL14 and WT1 associated protein (WTAP) on progressions of NPC. Finally, immunohistochemistry (IHC) and survival analysis were performed to confirm the correlation between METTL3, METTL14 and WTAP and NPC patients’ clinical outcomes.
Results: This study mapped m6A RNA modification and RNA expression profiles in normal nasopharynx, primary NPC, and recurrent NPC tissues. This study also explored the role of m6A modificators in NPC development and recurrence. METTL3, METTL14, and WTAP could promote invasion and metastasis of NPC, and that these three proteins could induce radiotherapy resistance in NPC cells through DNA repair. Moreover, we found that METTL3, METTL14, and WTAP promoted an increase in exosomes within NPC microenvironment.
Conclusions: This study suggests that the alteration of m6A modification in primary and recurrent NPCs may play an important role in the development and progression of NPC.
Keywords: Nasopharyngeal carcinoma; N6-methyladenosine; RNA modification; Recurrence; Occurrence
| Introduction | ▴Top |
Nasopharyngeal carcinoma (NPC) is a type of squamous head and neck cancer with variable geographic distributions and the highest incidence in Southeast Asia [1-3]. With the development of treatment techniques, the 5-year overall survival rate of NPC has reached 50-64%. However, 10-20% of patients with NPC still experience locoregional recurrence, metastasis, invasion, and radio resistance [4-6]. Therefore, determining biomarkers and potential mechanisms affecting carcinogenesis and recurrence is essential for developing a more personalized treatment for patients with NPC.
N6-methyladenosine (m6A) is a type of internal RNA modification in eukaryotes, which refers to the addition of a methyl group at position N6 of adenosine [7]. M6A modification is a reversible process regulated by modificators, including writers, erasers, and readers. With the development of high-throughput sequencing technologies and specific antibodies, m6A modifications have been studied in-depth; and m6A has important effects on various biological regulatory processes [8-11]. However, the m6A landscape in NPC, especially recurrent NPC, has not been reported, especially the differences between initial and recurrent NPCs compared to normal nasopharyngeal tissues, and their possible roles. Here, m6A RNA immunoprecipitation (RIP) sequencing and microarray sequencing were performed on 12 human tissue samples, including normal nasopharyngeal tissues, primary NPC tumor samples, and recurrent NPC tumor samples. The expression profiles of m6A-related and non-coding RNAs were constructed and explored. We also explored the role of m6A modificators in NPC development and recurrence.
| Materials and Methods | ▴Top |
Clinical sample and methylated RNA immunoprecipitation (meRIP) sequencing
In total, 24 NPC samples including paired primary and recurrent NPC sample were collected at the Xiangya Hospital of Central South University (Changsha, China). This study was approved by the Joint Ethics Committee of the Central South University Health Authority, and informed consent was obtained from all participants. Diagnoses of all specimens were confirmed via histopathological examination. Hybridization of cRNA to Arraystar Human mRNA and long noncoding RNA (lncRNA) epitranscriptomic microarray (8 × 60 K, Arraystar) was performed for sequencing; and the result was detected using an Agilent Scanner G2505C and analyzed using the Agilent Feature Extraction software (version 11.0.1.1). The study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
Public database
Public RNAseq data for cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) project (https://portals.broadinstitute.org/ccle) and Genomics of Drug Sensitivity in Cancer (GDSC) project (https://www.cancerrxgene.org/). All expression data were processed to transcripts per million (TPM) by using Python. Expression data from CCLE was normalized by using ln(TPM+1). RNA-sequencing (RNA-seq) data of the gene expression are available for download at The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/).
Transwell assays
The migratory capacity of the cells was tested using a transwell chamber (Millipore Corporation, USA). Cells were suspended in serum-free RPMI-1640 medium, and approximately 30,000 cells were seeded into the top chamber. RPMI-1640 medium containing 10% fetal bovine serum (FBS) was added to the bottom chamber. After incubation at 37 °C for 48 h, the cells that did not pass through the pores of the top chambers were erased. The chambers were then fixed in 4% paraformaldehyde (PFA) for 30 min before staining with 1% crystal violet for 10 min. Cells at the bottom of the top chamber were observed and counted using an inverted microscope.
Wound healing assay
Transfected cells were cultured in six-well plates for 24 h, starved with serum-free RPMI-1640 for 12 h, and wounded with a sterilized pipette tip to make a straight scratch. After gentle washing with phosphate-buffered saline (PBS), the cells were cultured in serum-free RPMI-1640 medium. Images were taken using a Leica inverted microscope at 0, 6, 12, and 24 h post-wounding (at least three areas were randomly selected for imaging).
Cell culture and small interfering RNA (siRNA) transfection
HK1 and S18 cells were cultured in a medium (RPMI-1640, Gibco, USA) with 10% (v/v) FBS at 37 °C and 5% CO2. siRNAs were transfected using Lipofectamine RNAiMax (Invitrogen, USA) using a standard protocol 24 - 48 h before analysis. The siRNAs used in this study were methyltransferase (METTL)3 siRNA (siMETTL3), siMETTL14, and WT1 associated protein (WTAP) siRNA (siWTAP).
qRT-PCR
Total RNA was extracted from all samples using TRIzol reagent (Life Technologies, USA) and subjected to reverse transcription using the 5 × all-in-one kit (US Everbright Inc., China). The expression levels of target RNAs were measured using SYBR Green Master Mix (Yeasen, China) using a StepOnePlus Real-Time PCR System (Applied Biosystems, USA). GAPDH was used as an endogenous control. The fold changes were calculated using the 2-ΔΔCq method. The primers used were as follows: the METTL3 forward primer 5’-CGTACTACAGGATGATGGCTTTC-3’ and reverse primer 5’-TTTCATCTACCCGTTCATACCC-3’, METTL14 forward primer 5’-AGTGCCGACAGCATTGGTG-3’ and METTL14 reverse primer 5’-GGAGCAGAGGTATCATAGGAAGC-3’, and WTAP forward primer 5’-GCAACCAAGGAACAAGAGATG-3 and reverse primer 5’-CAAGTTGATCGCTGGGTCTAC-3’.
Immunohistochemistry and tissue staining evaluation
The processing steps before adding primary antibody to tissue sections were the same as those for immunofluorescence. Diluted primary antibody was added, and the sections were incubated overnight at 4 °C. Subsequently, the tissue sections were washed thrice with PBST (PBS containing 0.05% tween-20) water and incubated with the diluted secondary antibody for 1 h at room temperature. After washing thrice with PBS, the color was developed with 3,3’ diaminobenzidine tetrahydrochloride (DAB) for 1 - 10 min, and the degree of staining was controlled under the microscope. Hematoxylin staining was performed for 2 min at room temperature. They were later placed in xylene twice for transparency. Staining was scored according to the staining intensity and distribution of the stained cells. The distribution was evaluated as none (0), ≤ 10% (1), 10-50% (2), 50-80% (3), and > 80% (4). Intensity was evaluated as none (0), faint (1), moderate (2), strong (3), or very strong (4). The antibodies included METTL3 and METTL14.
Tissue immunohistochemistry staining and microscopy
Tissue sections were dewaxed by immersion in ethanol solutions of different gradients for 5 min and then washed thrice with PBS. Citric acid buffer (pH: 6) was used for antigen repair and then washed twice. They were blocked with 5% bovine serum albumin (BSA, SIGMA, A-7030) in PBS for 1 h at room temperature. Primary antibodies were diluted in blocking buffer and incubated with tissue sections overnight at 4 °C. The tissue sections were then washed thrice with 0.05% PBST and incubated with secondary antibodies for 1 h at room temperature. Finally, they were washed thrice with 0.05% PBST and optionally stained with 4’,6-diamidino-2-phenylindole (DAPI, 1:1,000 in PBS). The primary antibodies used for immunoassays were METTL3 (CST, #86132), WTAP (CST, #41934), and METTL14 (CST, #51104).
Immunofluorescence staining and microscopy
Cells were seeded in a 35 mm glass-bottom dish (MatTek, P35GC-1.5-14-C). After the transfection and treatment of indicated dose of irradiation through a Precision X-ray machine (PXi, X-RAD 225 Lite), cells were firstly rinsed with PBS (BE17-516F) and fixed in 4% PFA (Affymetrix, 19943 1 LT) for 15 min at room temperature. Then they were washed three times by PBS and followed by permeabilization using 0.2% Triton X-100 in PBS for 15 min and washed three times by PBS. Then they were blocked by 5% BSA (SIGMA, A-7030) in PBS for 1 h at room temperature. Primary antibodies were diluted in blocking buffer and added for overnight incubation at 4 °C. After overnight incubation, the cells were washed three times with 0.05% PBST and incubated with secondary antibodies for 1 h at room temperature, including Alexa Fluor 405/488/594 goat anti-mouse/rabbit IgG conjugate (Abcam, 1:3,000). Finally, they were washed three times by 0.05% PBST and optionally stained with DAPI (1:1,000 in PBS) for 5 min at room temperature. The primary antibodies for immunoassays are METTL3 (CST, #86132), WTAP (CST, # 41934), and METTL14 (CST, #51104).
Exosome isolation
Exosomes were purified from NPC cell cultured media by exosome isolation kit methods. After 48 h of culture, conditioned medium containing 2% exosome-depleted FBS was collected. Recipient cells (2 × 105) were incubated with 2 µg exosomes for 48 h. The collected biofluid was centrifuged (Beckman Coulter, Brea, CA, USA) at 4,000 × g for 10 min at 4 °C to remove cell debris and then at 17,000 × g at 4 °C for 60 min to remove remaining macropolymers. The supernatant was passed through a 0.22-µm filter (Millipore, MA, USA) and further ultracentrifuged at 200,000 × g at 4 °C for 60 min to collect vesicles smaller than 100 nm and protein aggregates. The pellets were resuspended in PBS and ultracentrifuged at 200,000 × g for 60 min at 4 °C to eliminate contaminant proteins and then dissolved in ice-cold PBS for further analysis. Regarding the exosome isolation kit method, the collected biofluid was centrifuged at 4,000 × g for 10 min at 4 °C to remove cell debris and then passed through a 0.22-µm filter. After centrifugation at 3,000 × g for 30 min at 4 °C in the dialysis tube (Merck KGaA, Darmstadt, Germany), the supernatant was incubated with ExoQuick-TC™ exosome precipitation solution (YEXO96C-H, YISIKE bio, China) for 30 min (sera) or 6 h - overnight (media) at 4 °C. The amount of exosomes was measured using the enzyme-linked immunoassay (ELISA) kit (Novagen, Merck Group, Madison, USA).
Western blotting (WB)
For WB analysis, samples were boiled at 95 °C for 5 - 8 min in SDS loading buffer. Then they were subjected to electrophoresis in 8-12% SDS-polyacrylamide gels and transferred to the polyvinylidene difluoride membrane. The membranes were blocked with 5% non-fat milk in PBS for 1 h before being incubated with the primary antibody at 4 °C overnight. The primary antibodies for WB used in this study are CD9, CD63 and TSG101. Then the cells were washed three to four times with 0.1% PBST and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000) for 1 h at room temperature. The membranes were washed in 0.1% PBST four times before exposure. Chemiluminescent HRP substrate was purchased from Abcam (Catalog#: WBKLS0500). Images were acquired in a BIO-RAD Universal Hood II machine with ImageLab software.
Statistical analysis
The data were presented as mean ± standard deviation (SD) from technical triplicates. Comparisons between each group were calculated using Student’s t-test, two-tailed Fisher’s exact test method of summing small P values, one-way and two-way analysis of variance, and Bonferroni’s multiple comparison test as appropriate. A value of P < 0.05 was considered significant. GraphPad Prism version 7 was used for graphics (GraphPad Software, San Diego, CA, USA).
| Results | ▴Top |
Landscape of RNA m6A and clinical characteristics of NPC samples
To detect the alteration of m6A modification in carcinogenesis and recurrence of NPC, we collected specimens from normal nasopharyngeal tissues of patients with initial and recurrent NPC at Xiangya Hospital Central South University and detected alterations in intracellular m6A modification by two methods: m6A microarray and meRIP sequencing. Subsequently, we explored the alteration of m6A modification in nasopharyngeal carcinogenesis and recurrence by combining clinical data. We further examined the function of the writer molecules of m6A in nasopharyngeal carcinogenesis and progression using in vivo experiments (Fig. 1a, b) (Supplementary Material 1, www.wjon.org).
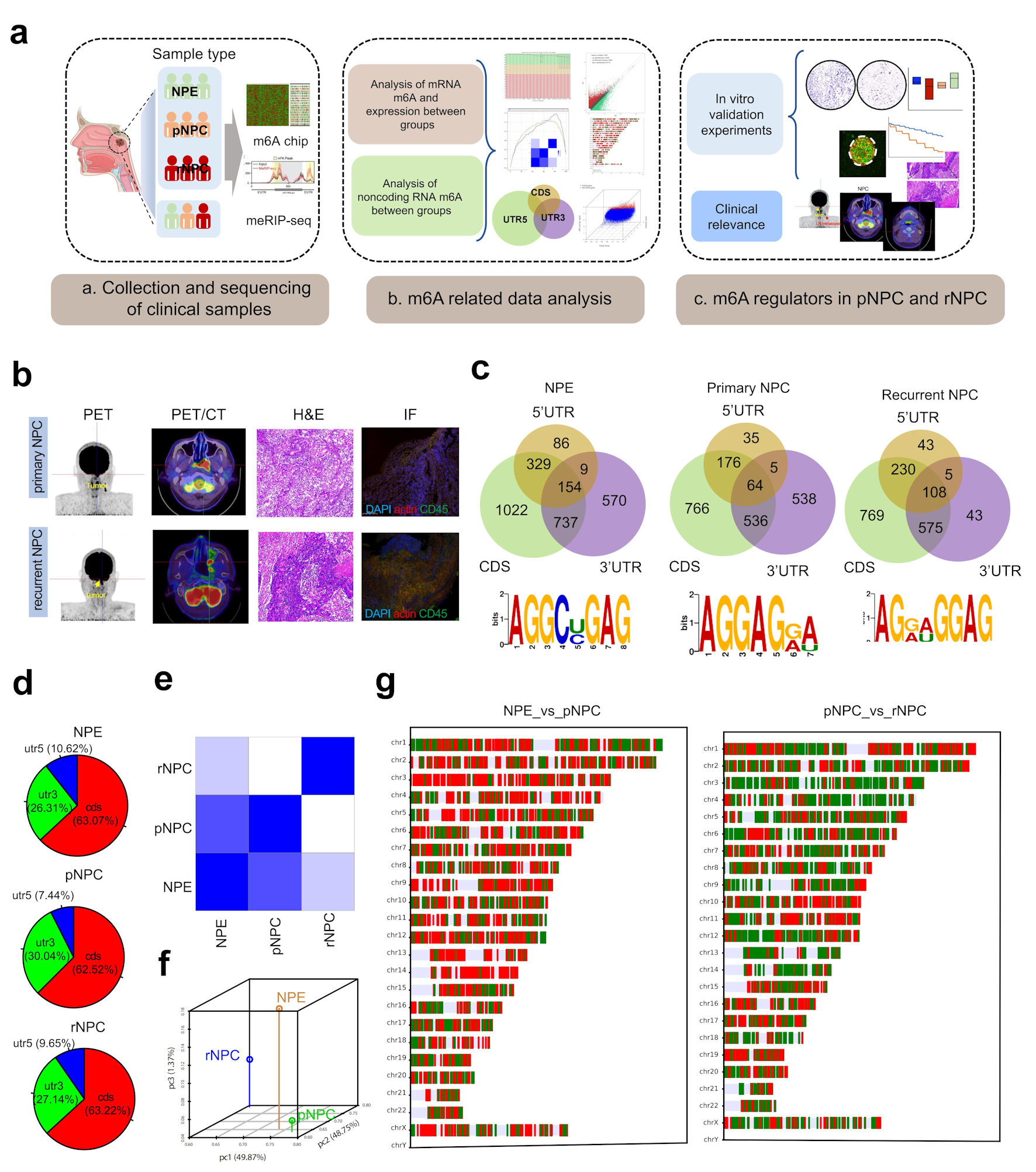 Click for large image | Figure 1. Landscape of RNA m6A modification in NPE, primary and recurrent nasopharyngeal carcinoma. (a) The experimental scheme for discovering and validating m6A modification in NPC. (b) PET/CT and immunohistochemistry (IHC) of representative primary and recurrent NPC patient. (c) The distribution of m6A on mRNA region and the m6A-prone motif in NPC tissues. (d) The distribution of differently methylated mRNA between groups. (e) Heatmap of PCA correlation between NPE, pNPC and rNPC mRNA expression. (f) Dot plot of PCA correlation between NPE, pNPC, and rNPC mRNA expression. (g) Distribution of differentially expressed mRNA molecules on chromosomes; red represents higher expression, blue represents lower repression. CDS: coding sequence; UTR: untranslated region; NPC: nasopharyngeal carcinoma; NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma; PET/CT: positron emission tomography/computed tomography scan; H&E: hematoxylin and eosin; m6A: N6-methyladenosine; IF: immunofluorescence. |
By microarray and meRIP sequencing, m6A modification was significantly more altered in initial and recurrent NPC samples compared to normal nasopharyngeal tissue. Intracellularly, normal nasopharyngeal epithelium (NPE) genes underwent m6A modification with 63.07%, 10.62%, and 26.31% occurring in the 3’ untranslated region (UTR), 5’UTR, and coding sequence (CDS) regions, respectively. In primary NPC tissue transcripts with m6A, 7.44%, 30.04%, and 62.52% modification occurred in the 3’UTR, 5’UTR, and CDS regions, respectively. In the recurrent NPC transcripts with m6A modification, 9.65%, 27.14%, and 63.22% occurred in the 3’UTR, 5’UTR, and CDS regions, respectively (Fig. 1c, d) (Supplementary Material 2, www.wjon.org).
To further detect intracellular mRNA expression alterations, we examined the expression of mRNA in the three different tissue types and obtained mRNA expression profiles in NPE and initial and recurrent NPCs (Fig. 1e-g) (Supplementary Materials 3, 4, www.wjon.org).
m6A-modified transcripts are significantly altered in NPE, primary NPC, and recurrent NPC
Analysis of differential m6A modification mRNA in normal NPE and initial and recurrent NPC samples showed that there were 1,465 mRNA transcripts with significantly higher m6A modification in primary NPC than in normal tissues. One hundred sixty-eight mRNA molecules displayed a significant reduction. In recurrent NPC, 1,613 mRNA molecules showed a significant increase in m6A modification (Fig. 2a). The top 1,000 molecules with the highest m6A levels were counted, and the results suggested that a larger number of molecules had equally different mRNA levels (Fig. 2b). To further investigate mRNA regulation by m6A modification of mRNA, the correlation between mRNA expression and m6A levels was examined in combination with mRNA expression in tissues. The results showed that, compared to NPE, there were primary NPC mRNAs whose expression was regulated by m6A and mRNAs in recurrent NPC. There was no region specificity in the distribution of mRNAs with m6A differences on chromosomes (Fig. 2c). Further analysis of the differentially expressed mRNAs showed that most of the differentially expressed mRNA molecules were regulated by m6A levels (Fig. 2d).
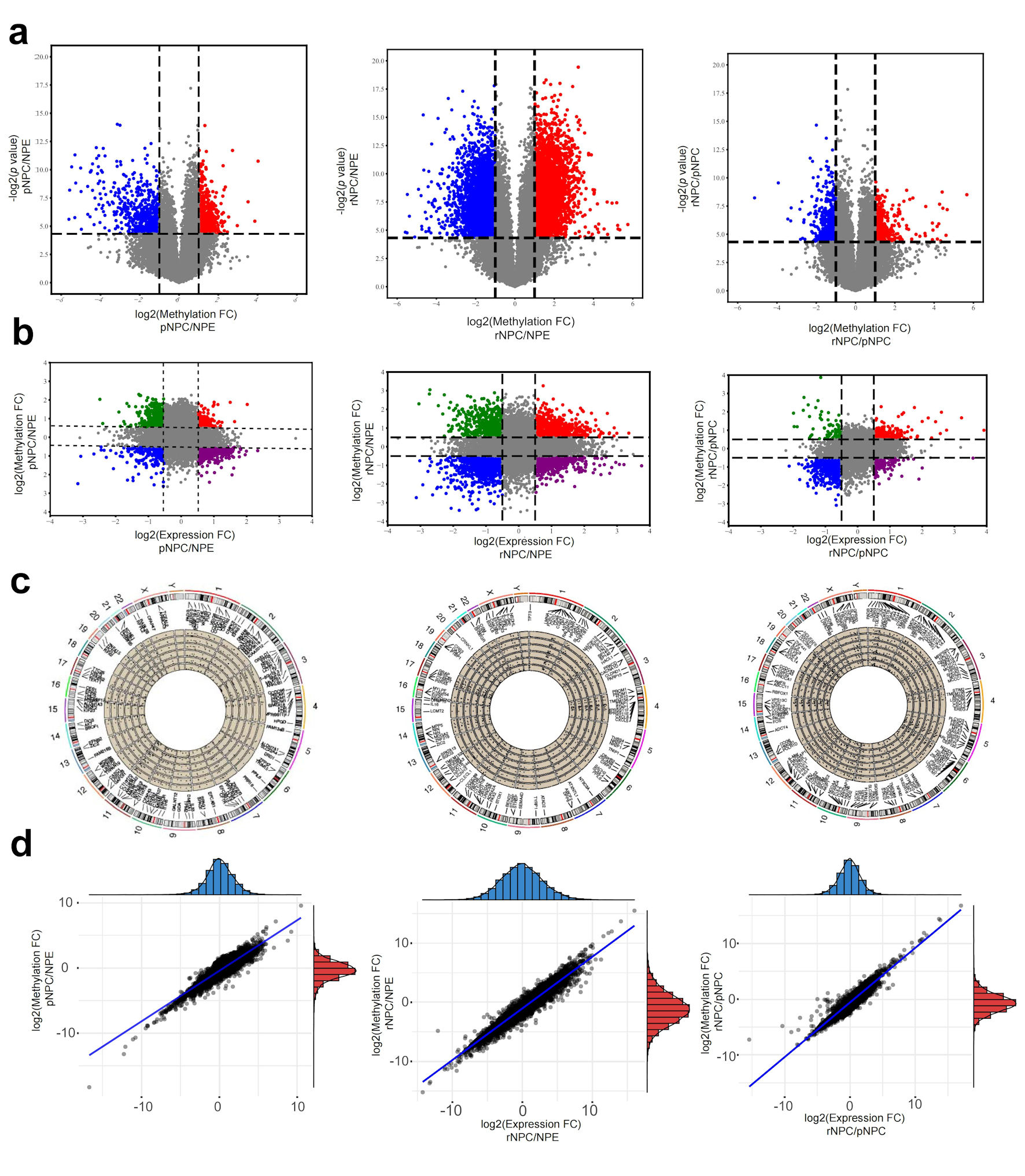 Click for large image | Figure 2. Differentially m6A modification and differentially expressed mRNAs in NPC. (a) Differential m6A modificated mRNA molecules in pNPC/NPE, rNPC/NPE and rNPC/pNPC groups (from left to right). (b) Relationship between mRNA methylation fold change and expression fold change in pNPC/NPE, rNPC/NPE and rNPC/pNPC groups (from left to right). (c) Distribution of differential mRNA molecules on chromosomes in pNPC/NPE, rNPC/NPE, and rNPC/pNPC groups. (d) Relationship between mRNA methylation and expression level in pNPC/NPE, rNPC/NPE, and rNPC/pNPC groups (from left to right). NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma. |
By GSEA pathways enriching the differentially expressed molecules, we found that compared to normal NPE, the differential molecules in primary NPC were mainly concentrated in cancer development-related signaling pathways, such as immunity change and metabolic reprogramming (Fig. 3a) (Supplementary Material 5, www.wjon.org). In contrast, the differential genes in recurrent NPC compared to primary NPC were focused on some interesting signaling pathways, such as cAMP-cGAS which may affect both radiotherapy and immunotherapy (Fig. 3b, c). These results may provide new ideas for the study of NPC carcinogenesis. Moreover, they initially suggested that recurrent NPC has different characteristics from the primary NPC. The visualization of meRIP sequencing of representative molecules showed that m6A in the 5’UTR and CDS regions were different. Moreover, 187 molecules showed significant differences in m6A modifications in both primary and recurrent NPCs.
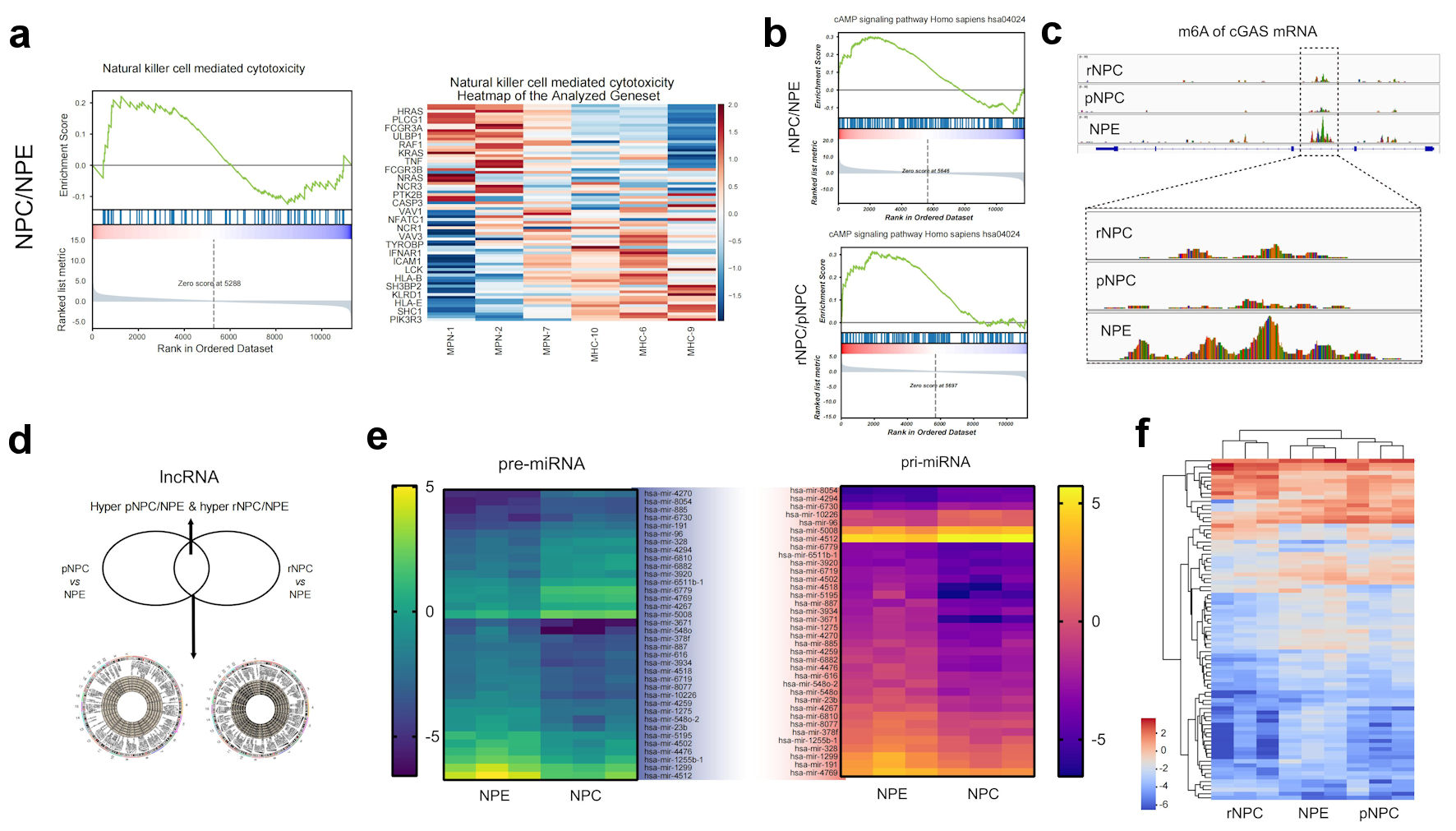 Click for large image | Figure 3. Function analysis of differently methylated mRNA and noncoding RNAs. (a) Represent GSEA enrichment function for differential methylation mRNA molecules in NPC. (b) Represent GSEA enrichment signaling pathways for differential methylation mRNA molecules in NPC. (c) m6A modification details of representative molecule cGAS by meRIP sequencing. (d) Differently methylated lncRNAs in primary NPC and NPE tissues. (e) Heatmap of differently methylated pre-miRNAs and pri-miRNAs in primary NPC and NPE tissues. (f) Heatmap of differently methylated snoRNA in NPC and NPE tissues. NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma; m6A: N6-methyladenosine; lncRNA: long noncoding RNA; me RIP: methylated RNA immunoprecipitation; cGAS: cyclic GMP-AMP synthase; miRNAs: micro RNAs; pre-miRNAs: precursor miRNAs; pri-miRNAs: primary miRNAs. |
In addition to mRNA, lncRNA, preRNA, and other non-coding RNAs have played important roles in the development and progression of various tumors, including NPC, in recent years [11-13]. To explore the possible roles of differentially m6A-modified lncRNAs during NPC carcinogenesis and recurrence, 1,167 m6A-associated lncRNAs were selected, which were hypermethylated in primary NPC/NPE and hypomethylated in primary NPC/NPE. Only 534 m6A lncRNAs were hypomethylated in recurrent NPC/primary NPC and hypermethylated in recurrent NPC/primary NPC (Fig. 3d). Furthermore, our results indicate that m6A-modified lncRNAs may regulate the expression of the numerous upregulated mRNAs associated with several significantly upregulated pathways, including the immune system and signal transduction processes (Supplementary Material 6, www.wjon.org).
In addition to lncRNA, we also examined the difference in m6A levels of preRNA and priRNA, as the precursor of miRNA, with 176 preRNAs and 134 priRNAs showing different m6A methylation. In the recurrent NPC/NPE comparison, 156 preRNAs and 78 priRNAs showed different m6A methylation levels. In the primary NPC/recurrent NPC comparison, 57 preRNA and 63 priRNAs had different m6a levels compared to recurrent NPC. These results suggested that m6A-induced miRNA splicing and processing may also play an important role in the process of NPC recurrence (Fig. 3e).
Furthermore, we examined the m6A levels of snoRNAs, and the results showed that there were 99 snoRNAs with altered m6A levels (59, upregulated; 40, downregulated) (Fig. 3f). As an RNA that has been gaining attention in recent years, snoRNAs play important roles in numerous biological processes.
m6A writers METTL3, METTL14, and WTAP anticipate NPC progression
As major reasons for treatment failure in NPC, the malignant phenotype of NPC includes mainly resistance and metastasis, affecting the tumor microenvironment through exosomes. The malignant phenotype of NPC mainly includes resistance to radiotherapy and the ability to invade and migrate. We subsequently knocked down the expression of these three writers using siRNA to knockdown NPC cells and showed that knockdown of METTL3, METTL14, and WTAP suppressed the invasive and migratory ability of NPC cells (Fig. 4a-c) (Supplementary Material 7, www.wjon.org).
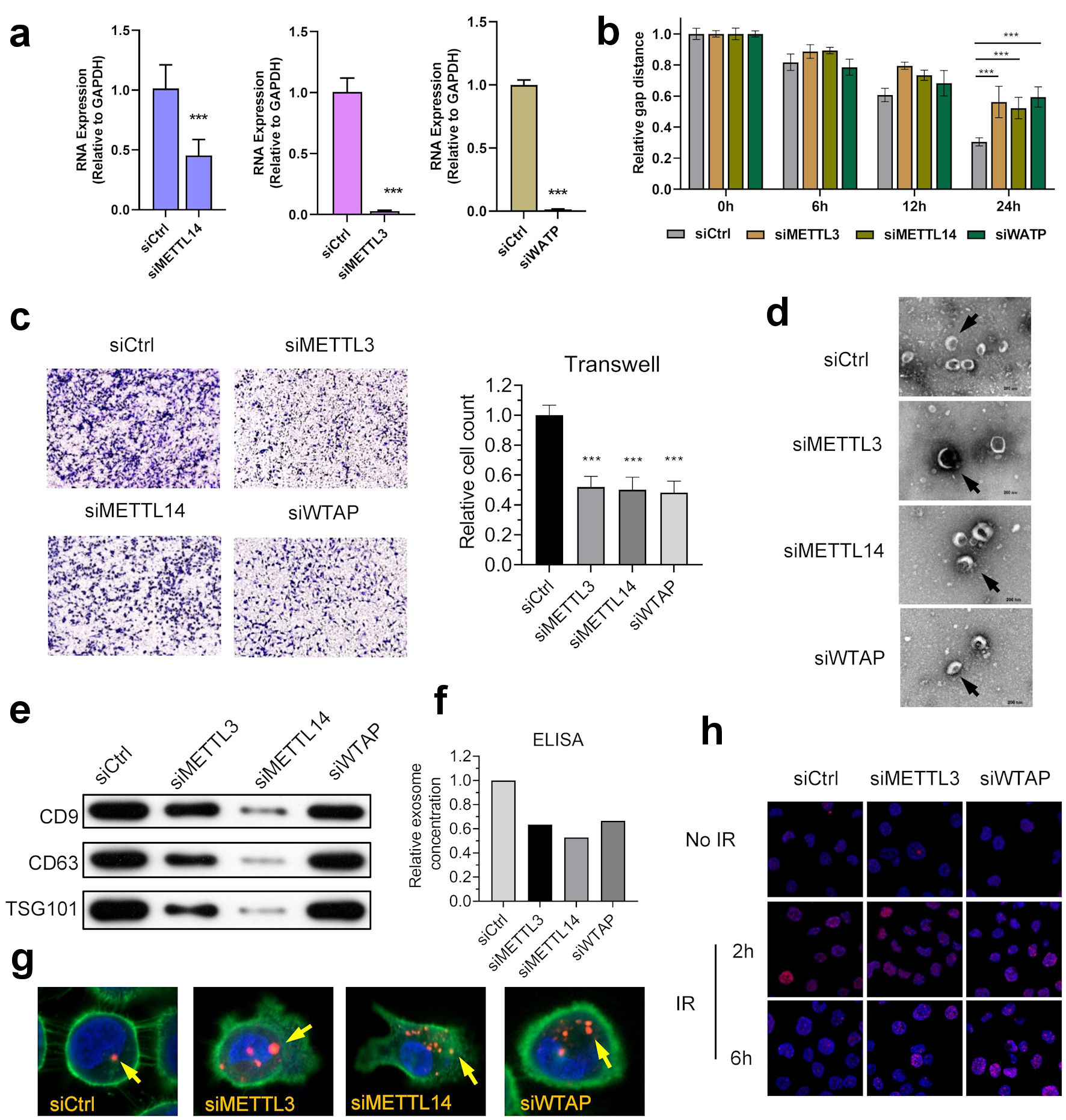 Click for large image | Figure 4. m6A regulators anticipate in the development and progression of NPC. (a) Knocking down of WTAP, METTL3 and METTL14 in NPC cell line HK-1. (b) WTAP, METTL3, and METTL14 promoted the migration ability of HNE2 cells transfected with siRNA assessed using by wound-healing assay. Images were acquired at 0 h to 24 h. Data have been represented as represent with mean ± SD. *P < 0.05; ***P < 0.001. (c) WTAP, METTL3, and METTL14 promoted the invasive ability of S18 cells after knockdown, as assessed using by Transwell® invasion assays. Images were acquired at 24 h and are representative of and presented from three independent experiments. Data have been represented as represent with mean ± SD. **P < 0.01; ***P < 0.001. (d) Representative cryo-electron microscopy images of exosomes in four groups (scale bar = 200 nm). (e) Western blotting analysis of exosome markers CD9, CD63, and TSG101 in four groups. (f) ELISA showed that knockdown of m6A writers would reduce exosome in NPC microenvironment. (g) Immunofluorescence showed that knockdown of m6A writers can induce uptake of exosome in NPC cells. (h) Represent image of γH2AX IRIF in 5-8F cells were quantified after 2 Gy IR at indicated recovery time in siControl, siMETTL3 and siMETTL14 group. METTL3: methyltransferase 3; METTL14: methyltransferase 14; WTAP: WT1 associated protein; ELISA: enzyme-linked immunoassay; IR: ionizing radiation; IRIF: IR-induced foci. |
We then examined the effects of METTL3, METTL14, and WTAP on microenvironment exosomes in NPC cells. Western blot and ELISA assay results showed a significant decrease in exosomes in NPC cell cultures after knockdown of METTL3, METTL14, and WTAP (Fig. 4d). Western blot and ELISA results showed a significant decrease in exosomes in NPC cell cultures after knockdown of METTL3, METTL14, and WTAP (Fig. 4e, f). The exosome uptake assay showed a significant increase in exosome uptake by NPC cells (Fig. 4g). These results suggest that METTL3, METTL14, and WTAP play roles in the development of NPC.
To further investigate the effect of the three molecules on the radiosensitivity of NPC, subsequent ionizing radiation-induced foci (IRIF) experiments showed that knockdown of METTL3 and WTAP significantly slowed down the clearance rate of H2AX, suggesting that the DNA repair rate of NPC cells was significantly slowed down (Fig. 4h).
Clinical relevance of m6A regulators in initial and recurrent NPCs
m6A is generated by the modification of the RNA molecule by a molecule complex, including writers, erasers, and readers. METTL3, METTL14, and WTAP are the core of the writer complex [7]. We then examined the possibility of METTL3, METTL14, and WTAP as molecular markers for predicting tumor prognosis in NPC. By immunohistochemistry, we found that the expressions of METTL3, METTL14, and WTAP were significantly higher in initial and recurrent NPC tissues than those in normal nasopharyngeal epithelial tissues (Fig. 5a, b). This is also consistent with the possible significant upregulation of m6A modifications in nasopharyngeal cancer tissues. Surprisingly, the expressions of METTL3, METTL14, and WTAP in recurrent NPC tissues were significantly lower than those in primary NPC tissues (Fig. 5b) (Supplementary Material 8, www.wjon.org). This suggests that the functions of METTL3, METTL14, and WTAP in the biological process of NPC recurrence is unexplored and should be further investigated. A combination of clinical samples from patients with NPC and survival analysis showed that patients with NPC with high expression of these three molecules had a significantly shorter survival time than patients with low expression (Fig. 5c). The results combined with the patient survival data showed that high expressions of METTL3, METLL14, and WTAP in NPC could be used as a prognostic marker. We further examined the correlation between writers, erasers, readers, and drugs in m6A in head and neck tumors using the IC50 values of cell lines and drugs using CCLE and GDSC. The results showed that m6A regulators in head and neck cell lines were positively correlated with numerous chemotherapy drugs like DNA damage-related drug (Fig. 5d) (Supplementary Materials 8, 9, www.wjon.org). suggesting that the related regulatory molecules including writers, erasers, and readers of m6A may serve as potential therapeutic targets for head and neck tumors, such as NPC, and that m6A may play an important role in drug resistance in tumors. We also obtained the expressions of writers and readers, such as METTL3, METTL14, and WTAP, in HNSCC from the TCGA database and assessed their correlation with survival. Results also showed that these core molecules were significantly correlated with poor survival in head and neck cancer patients. These results suggest an association between METTL3, and WTAP and nasopharyngeal carcinogenesis and recurrence (Fig. 5e).
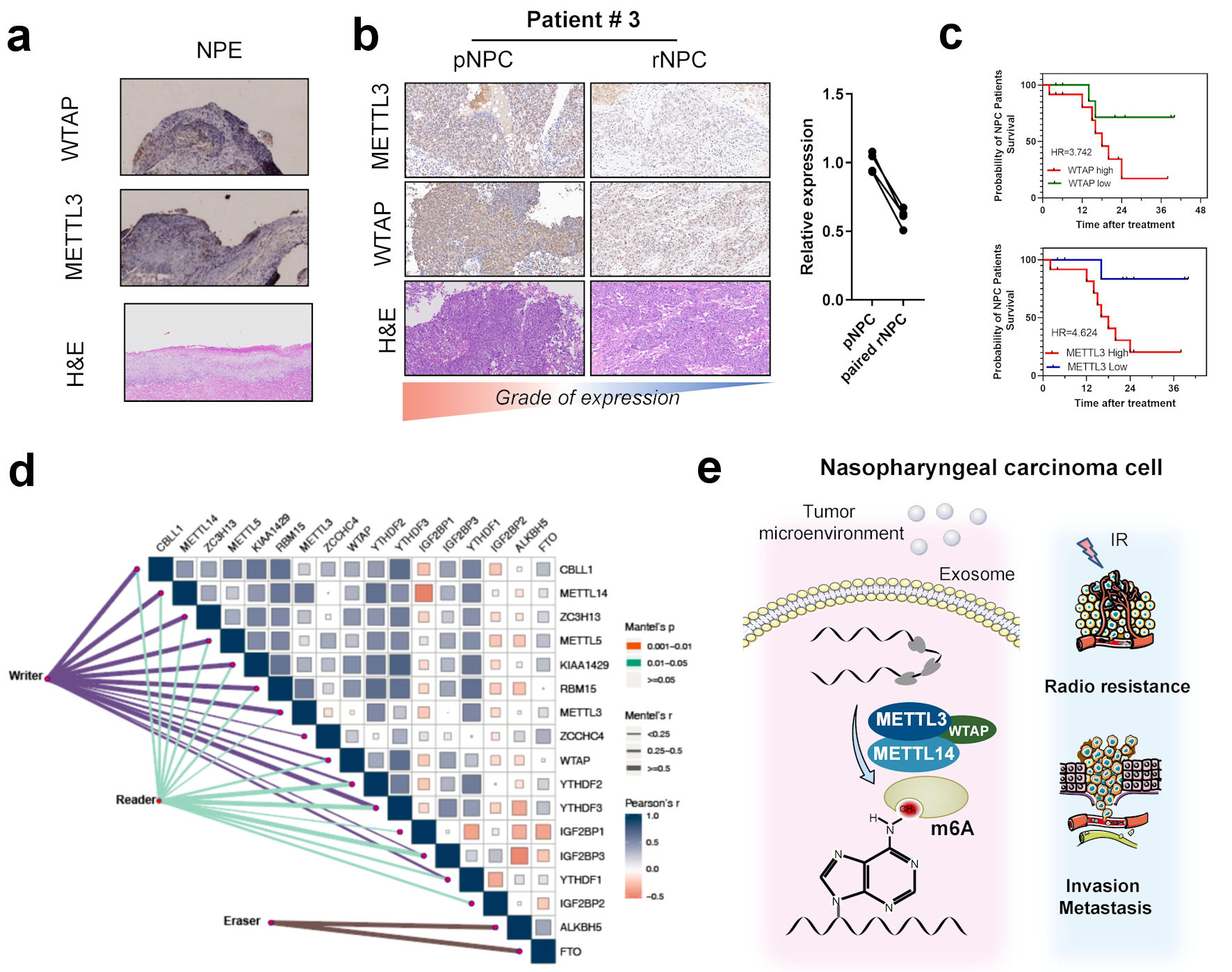 Click for large image | Figure 5. Correlation between m6A regulators with clinical outcomes in nasopharyngeal carcinoma. (a) Immunohistochemistry of m6A writer METTL3, WTAP, and hematoxylin and eosin (H&E) staining in NPE tissues. (b) Immunohistochemistry of m6A writer METTL3, WTAP, and H&E staining in primary NPC and paired recurrent NPC tissues. (c) K-M curve of overall survival in patients with different METTL3 and WTAP expression. (d) Correlation between expression of m6A regulators including eraser reader and writers in head and neck cancer patients’ tissues in TCGA-HNSC cohort. (e) The scheme of the role of m6A and regulators in the occurrence and recurrence of nasopharyngeal carcinoma. METTL3: methyltransferase 3; METTL14: methyltransferase 14; WTAP: WT1 associated protein; NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma; m6A: N6-methyladenosine; TCGA: The Cancer Genome Atlas; HNSC: head and neck squamous cell carcinoma. |
| Discussion | ▴Top |
In this research we examined m6A levels in recurrent NPC for the first time, obtained m6A modification profiles in normal nasopharyngeal tissues, primary and recurrent NPC, and compared the altered m6A modification levels in normal nasopharyngeal tissues, primary NPC and recurrent NPC tissues. The m6A regulator, especially the core molecules of m6A writer, METTL3, METTL14 and WTAP, were further tested experimentally in the development of nasopharyngeal carcinogenesis. As the component with the largest proportion of RNA modifications, m6A modification plays an important role in tumor development, but the occurrence and development of m6A in NPC are currently unclear.
The m6A modification of RNA has now been shown to anticipate in DNA damage repair of human cancer [14].
Insulin like growth factor binding protein 1 (IGFBP1), an important molecule of m6A, has also been shown to promote cell proliferation and metastasis in NPC driven by Myc [15], and WTAP promotes DIAPH1-AS1 can promote the proliferation and invasion of NPC [16]. METTL3 can also promote the development of NPC by promoting the stability of ZFAS1 [17]. circMAP3K4 promotes the development of NPC by regulating its ability to encode small peptides through regulating m6A modification [18]. Our results initially mapped RNA expression profiles and m6A modifications of RNA in NPE and initial and recurrent NPCs and preliminarily revealed the functions of m6A-related molecules in NPC development. Further mechanistic analysis is needed in future studies to explore the functions of m6A and related writers, erasers, and readers in NPC development.
The first choice of treatment for primary NPC is radiation therapy, but about 10% of patients will experience recurrence after treatment [19]. Recurrence has now become an important factor limiting the improvement of treatment outcome of NPC. However, the biological characteristics of recurrent NPC are still very poorly understood, and only a few studies have reported the involvement of molecules such as BRCA1 in recurrent nasopharyngeal carcinogenesis [20, 21]. Especially, the overall transcriptome mapping and epigenetic mapping of recurrent NPC have not been reported. In this study, we mapped the m6A RNA modification of recurrent NPC, which will also contribute to the subsequent in-depth study of recurrent NPC. In addition, our study showed that m6A regulators such as MRTTL3, METTL14 and WTAP were significantly downregulated after recurrence of NPC compared with before recurrence, which was a surprising and interesting finding. This suggests that recurrent NPC may have completely different biological characteristics from the primary NPC, and also suggests that recurrent NPC may require completely different treatment strategies from the primary NPC. More basic and clinical studies are needed to investigate these findings.
Conclusions
In this study, we mapped m6A RNA modification and RNA expression profiles in normal nasopharynx, primary NPC, and recurrent NPC tissues. We also initially examined the expressions of METTL3, METTL14, and WTAP in NPC and their effects on NPC development. Our results showed that METTL3, METTL14, and WTAP could promote invasion and metastasis of NPC, and that these three proteins could induce radiotherapy resistance in NPC cells through DNA repair. Moreover, we found that METTL3, METTL14, and WTAP promoted an increase in exosomes within NPC microenvironment. In conclusion, our study initially revealed the alteration of m6A modification in initial and recurrent NPCs and suggested that it may play an important role in the development and progression of NPC, providing new ideas for the study and potential therapeutic targets of NPC.
| Supplementary Material | ▴Top |
Suppl 1. Clinical characteristics of included patients.
Suppl 2. General information of m6A methylation in NPE and nasopharyngeal carcinoma samples.
Suppl 3. Differently m6A methylated mRNA in NPE, primary nasopharyngeal carcinoma, and recurrent nasopharyngeal carcinoma samples.
Suppl 4. The expression of mRNA in the three different tissue types and obtained mRNA expression profiles in normal nasopharyngeal epithelium and initial and recurrent NPCs.
Suppl 5. Enrichment of differently methylated mRNAs in NPE, primary NPC, and recurrent nasopharyngeal carcinoma samples.
Suppl 6. Possible cis-regulated genes.
Suppl 7. (A, B) m6A regulators participate in development and recurrence of nasopharyngeal carcinoma. (C, D) m6A regulators may participate in drug resistance of head and neck cancers.
Suppl 8. Expression of METTL3 and WTAP in primary NPC and paired recurrent nasopharyngeal carcinoma.
Suppl 9. m6A regulators in head and neck cell lines positively correlated with numerous chemotherapy drugs.
Acknowledgments
None to declare.
Financial Disclosure
This study was funded by the National Natural Science Foundation of China (82171118), Postdoctoral Innovative Talent Support Program of Hunan province (2021RC2017), the Natural Science Foundation of Hunan Province (2021JJ41027), Xiangya Hospital Funds for Young Scholar (2020Q13) and China Postdoctoral Science Foundation funded project (2021M693567, 2021TQ0374).
Conflict of Interest
The authors declare have no conflict of interest.
Informed Consent
Informed consent was obtained from all participants.
Author Contributions
Yu Min Wang and Zhou Ying Peng designed the project and completed the majority of experiments and analyzed the data. Ya Xuan Wang collected tissue samples. Lu Yuan Zhang made the bioinformatic analysis. Yu Min Wang and Zhou Ying Peng wrote the manuscript. Ruo Hao Fan and Hua Zhang revised the manuscript. Wei Hong Jiang is responsible for research supervision and funding acquisition. All authors read and approved the final manuscript.
Data Availability
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Abbreviations
CDS: coding sequence; UTR: untranslated region; FBS: fetal bovine serum; lncRNAs: long noncoding RNAs; m6A: N6-methyladenosine; NPC: nasopharyngeal carcinoma; NPE: normal nasopharyngeal epithelium; pNPC: primary nasopharyngeal carcinoma; rNPC: recurrent nasopharyngeal carcinoma; METTL3: methyltransferase 3; METTL14: methyltransferase 14; WTAP: WT1 associated protein; TCGA: The Cancer Genome Atlas; GSDC: genomics of drug sensitivity in cancer; CCLE: Cancer Cell Line Encyclopedia
| References | ▴Top |
- Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64-80.
doi - Liu YP, Wen YH, Tang J, Wei Y, You R, Zhu XL, Li J, et al. Endoscopic surgery compared with intensity-modulated radiotherapy in resectable locally recurrent nasopharyngeal carcinoma: a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22(3):381-390.
doi - Sun XS, Li XY, Chen QY, Tang LQ, Mai HQ. Future of Radiotherapy in Nasopharyngeal Carcinoma. Br J Radiol. 2019;92(1102):20190209.
doi pubmed - Wang Y, Peng Z, Wang Y, Yang Y, Fan R, Gao K, Zhang H, et al. Immune microenvironment change and involvement of circular RNAs in TIL cells of recurrent nasopharyngeal carcinoma. Front Cell Dev Biol. 2021;9:722224.
doi pubmed - Peng Z, Wang Y, Wang Y, Fan R, Gao K, Zhang H, Xie Z, et al. Preliminary efficacy report and prognosis analysis of endoscopic endonasal nasopharyngectomy for recurrent nasopharyngeal carcinoma. Front Surg. 2021;8:713926.
doi pubmed - Ma BBY, Lim WT, Goh BC, Hui EP, Lo KW, Pettinger A, Foster NR, et al. Antitumor activity of nivolumab in recurrent and metastatic nasopharyngeal carcinoma: an international, multicenter study of the Mayo Clinic phase 2 consortium (NCI-9742). J Clin Oncol. 2018;36(14):1412-1418.
doi pubmed - Liu J, Li K, Cai J, Zhang M, Zhang X, Xiong X, Meng H, et al. Landscape and regulation of m(6)A and m(6)Am methylome across human and mouse tissues. Mol Cell. 2020;77(2):426-440.e426.
doi pubmed - Tan F, Zhao M, Xiong F, Wang Y, Zhang S, Gong Z, Li X, et al. N6-methyladenosine-dependent signalling in cancer progression and insights into cancer therapies. J Exp Clin Cancer Res. 2021;40(1):146.
doi pubmed - Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, et al. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol Cancer. 2020;19(1):105.
doi pubmed - Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20(6):303-322.
doi pubmed - Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vagbo CB, Steiner FA, et al. Splice site m(6)A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell. 2021;184(12):3125-3142.e3125.
doi pubmed - Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37(3):270-288.
doi pubmed - Gu C, Shi X, Dai C, Shen F, Rocco G, Chen J, Huang Z, et al. RNA m(6)A modification in cancers: molecular mechanisms and potential clinical applications. Innovation (Camb). 2020;1(3):100066.
doi pubmed - Zhang C, Chen L, Peng D, Jiang A, He Y, Zeng Y, Xie C, et al. METTL3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of DSBs by Modulating DNA-RNA Hybrid Accumulation. Mol Cell. 2020;79(3):425-442.e427.
doi pubmed - Du M, Peng Y, Li Y, Sun W, Zhu H, Wu J, Zong D, et al. MYC-activated RNA N6-methyladenosine reader IGF2BP3 promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cell Death Discov. 2022;8(1):53.
doi pubmed - Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ, Lv JW, Zhang LL, et al. WTAP-mediated m(6)A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ. 2022;29(6):1137-1151.
doi pubmed - Peng J, Zheng H, Liu F, Wu Q, Liu S. The m6A methyltransferase METTL3 affects autophagy and progression of nasopharyngeal carcinoma by regulating the stability of lncRNA ZFAS1. Infect Agent Cancer. 2022;17(1):1.
doi pubmed - Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC, Liang H, Mei J, et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer. 2022;21(1):93.
doi pubmed - Peng Z, Wang Y, Fang Y, Wang Y, Yuan X, Shuai M, Xie S, et al. Salvage Endoscopic skull base surgery: another treatment option after immunotherapy for recurrent nasopharyngeal carcinoma. Front Immunol. 2022;13:899932.
doi pubmed - Cho WCS, Tse KP, Ngan RKC, Cheuk W, Ma VWS, Yang YT, Yip TTC, et al. Genomic characterization reveals potential biomarkers in nasopharyngeal carcinoma patients with relapse. Expert Rev Mol Diagn. 2020;20(11):1149-1159.
doi pubmed - He Y, Zhang L, Zhou R, Wang Y, Chen H. The role of DNA mismatch repair in immunotherapy of human cancer. Int J Biol Sci. 2022;18(7):2821-2832.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


