| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 2, April 2024, pages 181-191
Spinster Homologue 2 Expression Correlates With Improved Patient Survival in Hepatocellular Carcinoma Despite Association With Lymph-Angiogenesis
Joy Sarkara, h, Masanori Oshia, b, h, Vikas Satyanandaa, Kohei Chidaa, Li Yanc, Aparna Maitia, Nitai Haita, Itaru Endob, Kazuaki Takabea, b, d, e, f, g, i
aDepartment of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14263, USA
bDepartment of Gastroenterological Surgery, Yokohama, Kanagawa 236-004, Japan
cDepartment of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14263, USA
dDepartment of Surgery, University at Buffalo Jacobs School of Medicine and Biomedical Sciences, the State University of New York, Buffalo, NY, USA
eDepartment of Breast Surgery and Oncology, Tokyo Medical University, Tokyo 160-8402, Japan
fDepartment of Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata 951-8510, Japan
gDepartment of Breast Surgery, Fukushima Medical University, Fukushima, Japan
hThese authors contributed equally to this work.
iCorresponding Author: Kazuaki Takabe, Department of Surgical Oncology, Roswell Park Cancer Institute, Buffalo, NY 14263, USA
Manuscript submitted September 27, 2023, accepted January 30, 2024, published online March 21, 2024
Short title: SPNS2 Linked to Improved Survival in HCC
doi: https://doi.org/10.14740/wjon1732
| Abstract | ▴Top |
Background: Spinster homologue 2 (SPNS2) is a transporter of sphingosine-1-phosphate (S1P), a bioactive lipid linked to cancer progression. We studied the link between SPNS2 gene expression, tumor aggressiveness, and outcomes in patients with hepatocellular carcinoma (HCC).
Methods: Gene expression in patients with HCC was analyzed from the Cancer Genome Atlas (TCGA) (n = 350) and GSE76427 (n = 115) as a validation cohort, as well as liver tissue cohort GSE6764 (n = 75).
Results: High-SPNS2 HCC was significantly associated with high level of lymph-angiogenesis-related factors. SPNS2 expression was significantly higher in normal liver and early HCC versus advanced HCC (P < 0.02). High SPNS2 levels enriched immune response-related gene sets; inflammatory, interferon (IFN)-α, IFN-γ responses, and tumor necrosis factor (TNF)-α, interleukin (IL)-6/Janus kinase/signal transducer and activator of transcription (JAK/STAT3) signaling, complement and allograft rejection, but did not significantly infiltrate specific immune cells nor cytolytic activity score. High-SPNS2 HCC enriched tumor aggravating pathway gene sets such as KRAS (Kirsten rat sarcoma virus) signaling, but inversely correlated with Nottingham histological grade, MKI67 (marker of proliferation Ki-67) expression, and cell proliferation-related gene sets. Further, high-SPNS2 HCC had significantly high infiltration of stromal cells, showing that low-SPNS2 HCC is highly proliferative. Finally, high-SPNS2 HCC was associated with better disease-free, disease-specific, and overall survival (P = 0.031, 0.046, and 0.040, respectively).
Conclusions: Although SPNS2 expression correlated with lymph-angiogenesis and other cancer-promoting pathways, it also enriched immune response. SPNS2 levels were higher in normal liver compared to HCC, and inversely correlated with cancer cell proliferation and better survival. SPNS2 expression may be beneficial in HCC patients despite detrimental in-vitro effects.
Keywords: SPNS2; Sphingosine-1-phosphate; Hepatocellular carcinoma; Lymph-angiogenesis; Proliferation; Immune response; Gene set enrichment analysis
| Introduction | ▴Top |
Hepatocellular carcinoma (HCC) is the most common type of liver cancer, accounting for up to 90% of all primary liver cancers [1]. HCC most commonly arises from a background of liver injury, chronic inflammation, and cirrhosis [2]. Its bleak 18-20% 5-year survival [1, 3] is due in part to the facts that recurrence after resection of HCC is high at 50-70% at 5 years, and many patients present with locally advanced or metastatic disease precluding resection. Systemic treatment options for HCC have historically been limited [3], however, the landscape of systemic treatment is now evolving [4]. Thus, there is interest in identifying biomarkers for appropriate treatment selection and prognosis.
Sphingosine-1-phosphate (S1P) is a sphingolipid mediator which is produced in cells by the phosphorylation of sphingosine [5-7] and has been shown to play a role in inflammation-associated cancers [8], such as HCC. S1P must be transported out of cells in order to act on its receptors [9, 10], and therefore functions in an “inside-out” signaling mechanism [5] to regulate tumor cell growth and migration [11, 12], lymphangiogenesis [13, 14], angiogenesis [12], and recruitment of immune cells [15, 16]. Spinster homologue 2 (SPNS2) is an S1P transporter which was first discovered in 2009 [17] and was shown to play a role in lymphangiogenesis [9] and in inflammatory and autoimmune diseases [18]. SPNS2 has recently been linked to metastasis of HCC in the setting of iron deficiency [19], suggesting that its expression could be a therapeutic target in HCC.
The mechanisms by which SPNS2 regulate cancer physiology include the promotion of inflammation, modulation of cancer cell survival and migration, and modification of the tumor microenvironment (TME) [20]. These mechanisms are both linked to and independent of the concentration of S1P [20]. However, the net effect of SPNS2 on cancer progression is difficult to describe, due to the multiple complex and sometimes conflicting interactions within the TME. Therefore, the aim of our study was to evaluate the effect of SPNS2 expression in HCC on pro- and anti-cancer gene expression, immune cell recruitment, and clinical outcomes using a large transcriptomic database. Given the known effects of SPNS2 on lymphangiogenesis and cancer cell migration, we hypothesized that increased SPNS2 expression would correlate with higher HCC stage and poorer survival.
| Materials and Methods | ▴Top |
Clinical data acquisition
The clinicopathological and gene expression levels of patients with HCC from The Cancer Genome Atlas (TCGA) [21] was retrieved through cBioPortal as described previously [21-24]. The Gene Expression Omnibus (GEO) data set GSE76427 [25] was used as a validation cohort. A total of 473 patients with HCC were included in the analysis. GEO data set GSE6764 [26] was used to analyze gene expression profiles at various stages of liver fibrosis, cirrhosis and cancer. GSE6764 contained 75 patient samples with 13 samples from cirrhotic tissue, 17 dysplastic nodules, and 35 HCCs. Given that all the cohorts used in this study, TCGA and GEO datasets, are deidentified and publicly available, the Institutional Review Board approval was waived and informed consent was not applicable to this study. Additionally, ethical compliance is not applicable as no human or animal subjects were used in this study.
Gene set enrichment analysis (GSEA)
GSEA was performed using the publicly available software provided by the Broad Institute [27] as we have previously described [28-31]. Hallmark collection in the Molecular Signatures Database (MSigDB) was used for this study. Briefly, GSEA ranks all genes (usually around 200) in each gene set, then an enrichment score was calculated for each set. The enrichment score correlates with the frequency that members of that gene set occur at the top or bottom of the ranked data set, and therefore with expression of genes in each set. To this end, the value of the score is arbitrary and its worth is in its comparison. The statistical significance of GSEA was determined using the false discovery rate (FDR) of 0.25 throughout the study as recommended by the developer of the Broad Institute.
Immune cell composition and scores related to immune activity
As we previously reported [32-34], immune cell composition in a tumor was analyzed using xCell, a computational algorithm for enumerating cell subsets from the transcriptome reported by Aran et al in 2017 [35]. It integrates the deconvolution approaches used in CIBERSORT [36]. xCell algorithm estimates cell type fractions by comparing 489 gene signatures corresponding to 64 cell types, including lymphatic endothelial cells [37], microvascular endothelial cells [38], endothelial cells, pericytes and immune cells as we have previously reported [39-41].
Statistical analysis
All the statistical analyses were performed using R software [42]. Kaplan-Meier survival analysis was performed in R for the survival analysis. A P value of < 0.05 was considered statistically significant. One-way analysis of variance (ANOVA) was used to determine the significance of difference in various groups. We used Mann Whitney U test (two group comparison) and Kruskal test for multiple group comparison.
| Results | ▴Top |
SPNS2 expression levels correlate with lymphangiogenesis and angiogenesis
We first investigated whether SPNS2 expression in HCC tumors is associated with lymphangiogenesis-related gene expression. We found that SPNS2-high HCCs were significantly infiltrated by lymphatic endothelial cells (estimated using xCell algorithm) in both the TCGA (P < 0.01) and GSE76427 cohorts (P = 0.046) (Fig. 1a). Several markers of lymphangiogenesis (platelet endothelial cell adhesion molecule (PECAM)1, LYVE1, SOX 18, chemokine (C-X-C motif) ligand (CXCL)12, chemokine (C-X-C motif) receptor (CXCR)4, Prospero homeobox (PROX)1), were significantly upregulated in SPNS2-high tumors in both cohorts (all P < 0.006, except for CXCR4 and PROX1 in GSE76427) (Fig. 1b). We then conducted GSEA comparing high vs. low expression of SPNS2 in TCGA and GSE76427 cohorts and found that high expression of SPNS2 significantly enriched angiogenesis-related gene sets (Fig. 1c). Despite this gene set enrichment, SPNS2-high HCCs showed a significant infiltration of pericytes, and not microvascular endothelial cells, or endothelial cells (Fig. 1d).
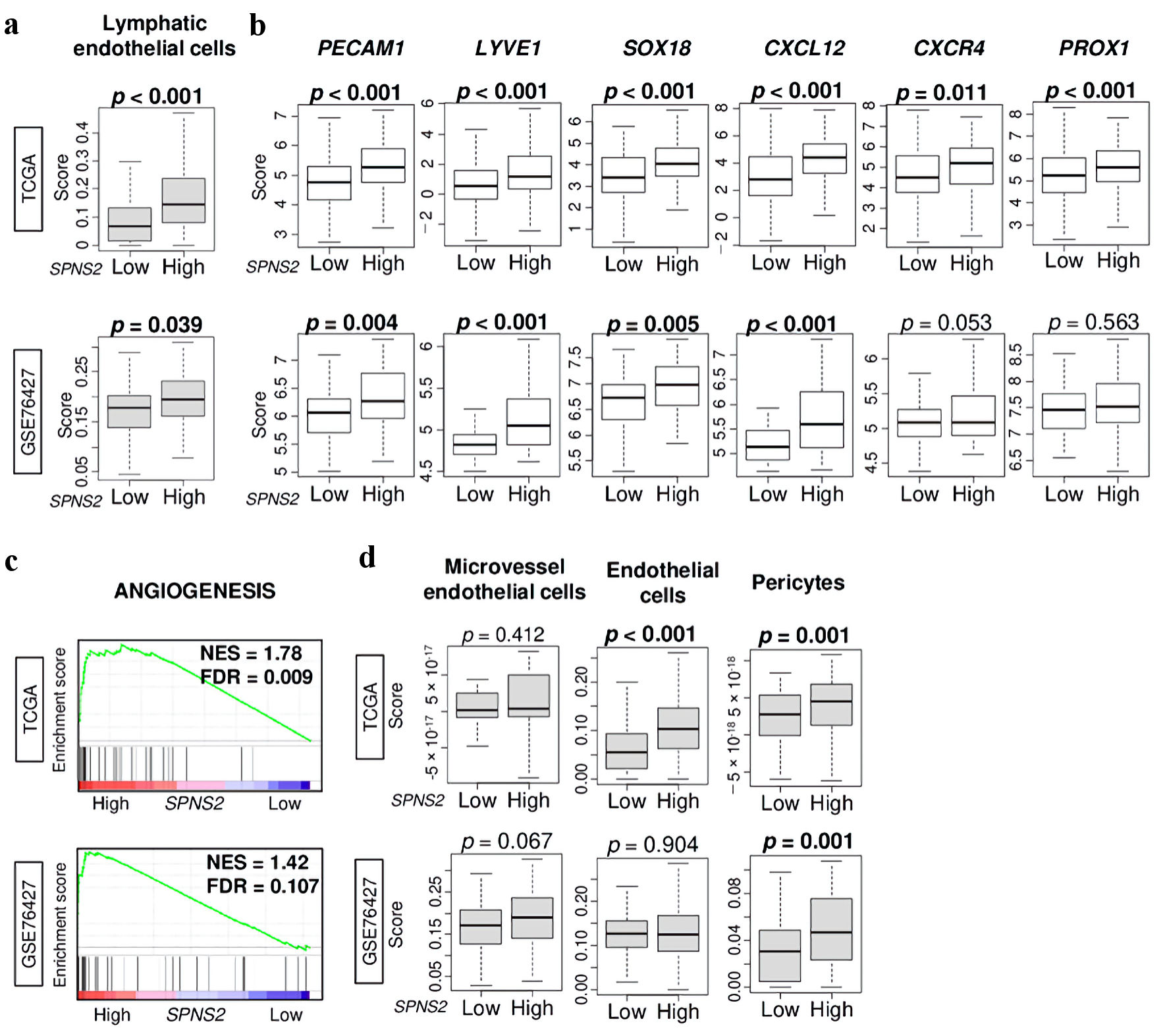 Click for large image | Figure 1. Lymphangiogenesis-related gene expressions by high vs. low SPNS2 expression in HCC of TCGA cohort. (a) Lymphatic endothelial cells estimated using xCell algorithm. (b) Gene expression levels of PECAM1, LYVE1, SOX18, CXCL12, CXCR4, and PROX1 in HCC of TCGA. (c) Gene set enrichment analysis (GSEA) of SPNS2 high HCC in two cohorts: angiogenesis-related pathway. The left side represents high expression, and the right side represents low expression of SPNS2. (d) Boxplots of microvascular endothelial cells, endothelial cells, and pericytes by low and high SPNS2 HCC estimated using xCell algorithm. CXCL: chemokine (C-X-C motif) ligand; CXCR: chemokine (C-X-C motif) receptor; HCC: hepatocellular carcinoma; PECAM: platelet endothelial cell adhesion molecule; PROX: Prospero homeobox; S1P: sphingosine-1-phosphate; SOX: Sry-type HMG box; SPNS2: spinster homologue 2; TCGA: The Cancer Genome Atlas. |
SPNS2 expression is inversely correlated with tumor grade and size
Given that SPNS2 expression was associated with lymphangiogenesis, we hypothesized that SPNS2 expression would also be associated with HCC tumor progression. To test this hypothesis, we compared SPNS2 expression in normal liver tissue and HCC tumors in the TCGA and GSE76427 cohorts. We found that in both cohorts, SPNS2 expression was actually higher in normal liver than in HCC tumors (P = 0.016 and P < 0.001 in TCGA and GSE76427, respectively) (Fig. 2a). In order to further query this unexpected finding, we evaluated the expression of both SPNS2 and lymphatic vessel endothelial hyaluronic acid receptor 1 (LYVE1) in the GSE6764 cohort which included a spectrum of liver tissues from normal liver tissue to very advanced HCC as described above. We found that the expression of SPNS2 was inversely correlated with degree of advancement of HCC, and that this was mirrored in expression of LYVE1 (both P < 0.001) (Fig. 2b). This demonstrated that although SPNS2 expression decreased in correlation with advancement of HCC, lymphangiogenesis similarly decreased, which may explain the above findings.
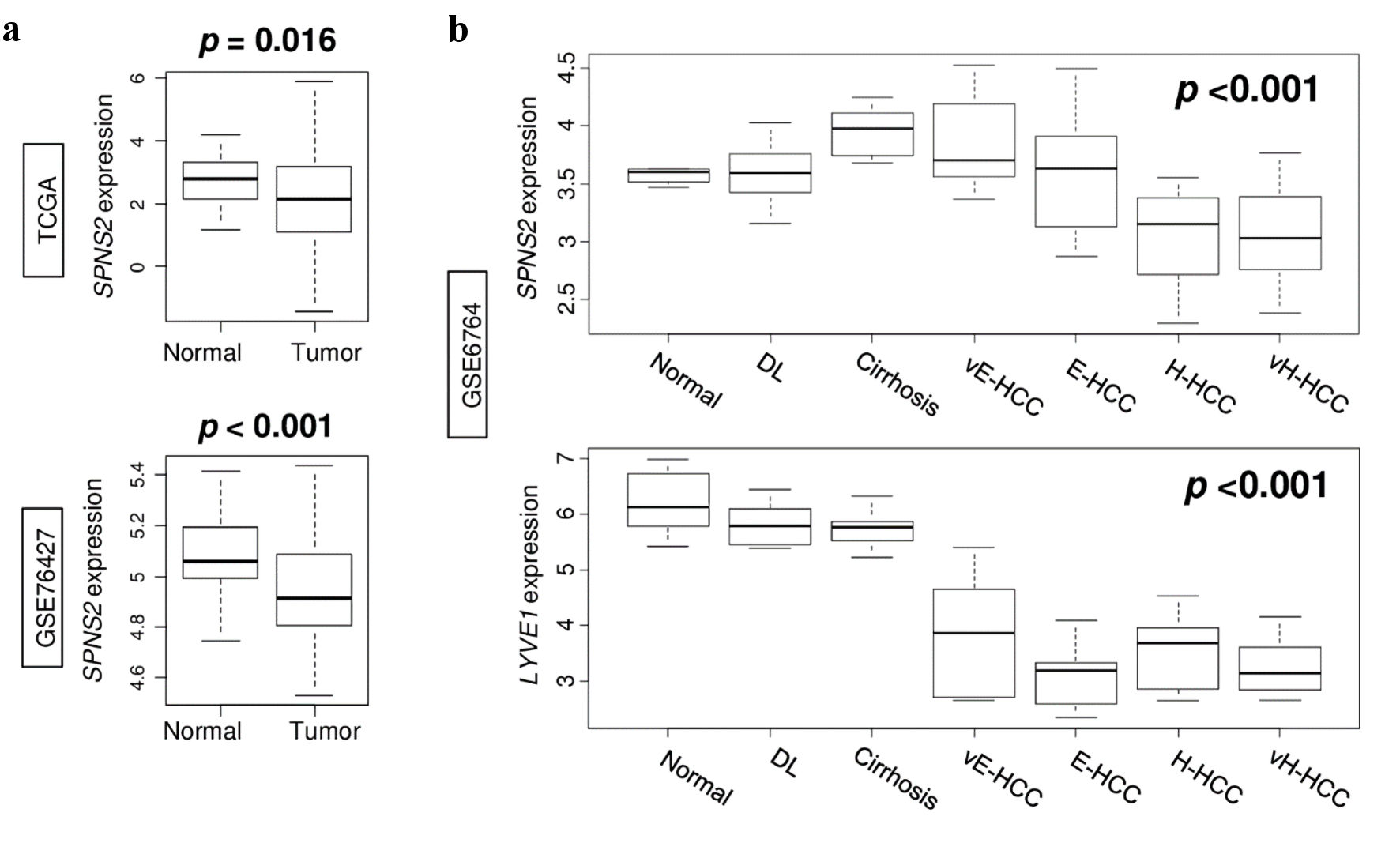 Click for large image | Figure 2. Gene expression in the spectrum of normal liver tissue to very advanced HCC. (a) SPNS2 expression in normal tissue compared to HCC tumors in TCGA and GSE76427 cohorts. (b) Expression of SPNS2 and lymphangiogenesis-related gene LYVE1 in GSE6764 cohort. vE-HCC: very early HCC; E-HCC: early HCC; H-HCC: advanced HCC; vH-HCC: very advanced HCC; HCC: hepatocellular carcinoma; TCGA: The Cancer Genome Atlas; SPNS2: spinster homologue 2; DL: dysplasia. |
SPNS2-high HCC enriches immune response pathways, but this does not correlate with consistent immune cell infiltration in the TME
Given the unexpected finding that SPNS2 expression was lower in HCC tumors compared to normal liver tissue, we conducted GSEA comparing high vs. low SPNS2 expression in HCC of TCGA and GSE76427 cohorts. We found that high SPNS2-expressing in HCC significantly enriched inflammation and immune response-related gene sets including inflammatory response, tumor necrosis factor (TNF)-α signaling, interleukin (IL)-6 signaling, interferon (IFN)-α response, IFN-γ response, allograft rejection, and complement (Fig. 3a). However, this did not translate to an increased host immune cell infiltration in the TME by xCell algorithm. SPNS2-high HCC did not demonstrate significant infiltration of lymphocytes or myeloid-derived immune cells (Fig. 3b), as would have been expected based on the GSEA. Furthermore, there was no consistent infiltration of pro-cancer immune cells such as regulatory T cells or M2 macrophages (Fig. 3c). There was no difference in overall cytolytic activity score (CAS) by SPNS2 expression (Fig. 3d).
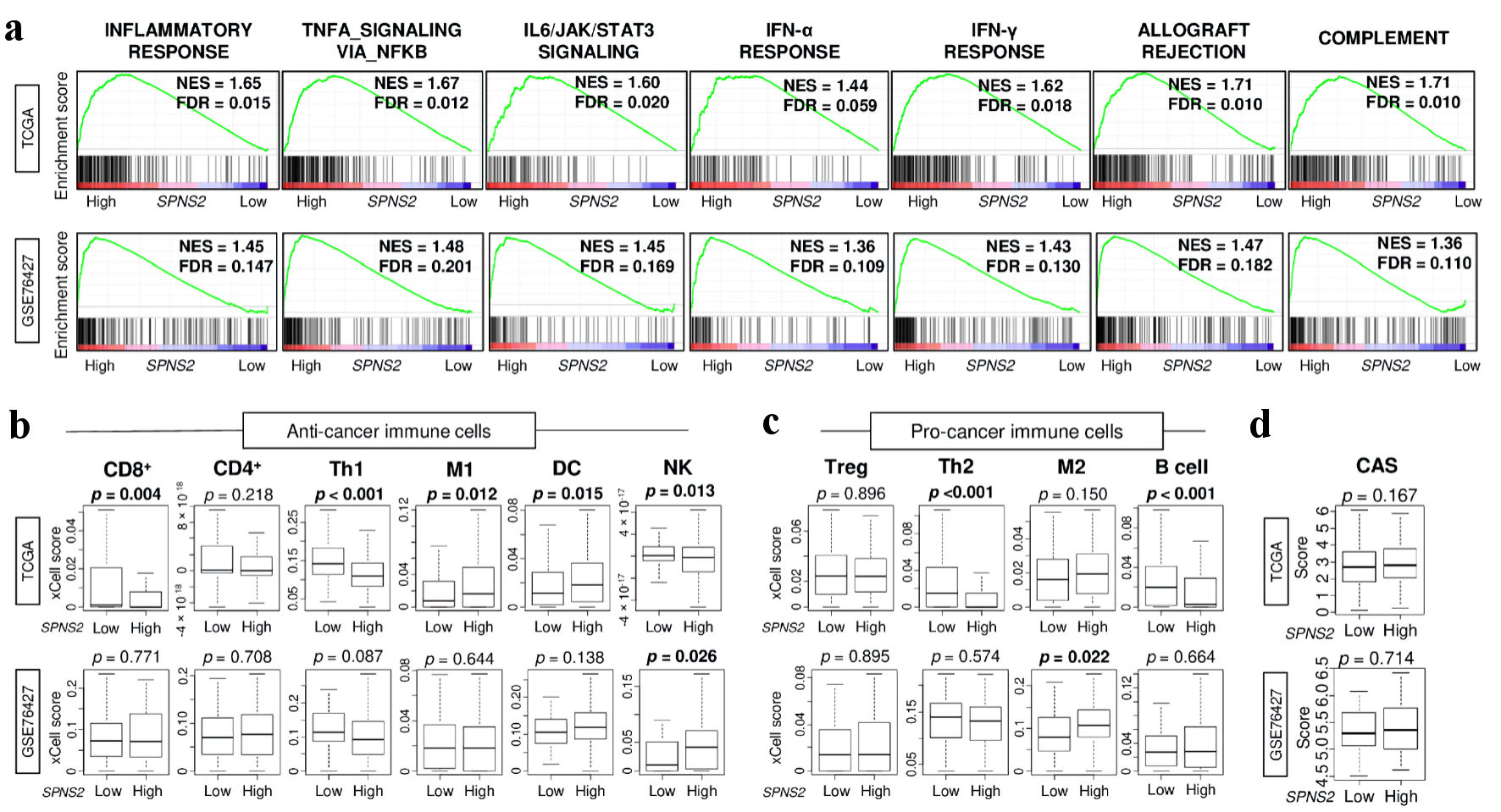 Click for large image | Figure 3. Immune response and immune cell infiltration in SPNS2-high HCC in two cohorts. (a) Gene set enrichment analysis (GSEA) of inflammation and immune response-related pathways: inflammatory response, TNF-α signaling, IL-6 signaling, IFN-α response, IFN-γ response, allograft rejection, and complement. The left side represents high expression, and the right side represents low expression of SPNS2. SPNS2 high and low expression groups were divided by median to perform the analysis. Normalized enrichment score (NES) and false discovery rate (FDR) of each analysis are shown in the graph. FDR of less than 0.25 were defined as statistically significant following the recommendation of the developer. (b) Estimated abundance of anti-cancer immune cells using xCell algorithm on each transcriptome (c) Estimated abundance of pro-cancer immune cells. (d) Cytolytic activity score (CAS). CD8: CD8 T cells; CD4: CD4 T cells; Th1: type 1 helper T cells; M1: M1 macrophage; DC: dendritic cells; NK: natural killer cells; Treg: regulatory T cells; Th2: type 2 helper T cells; M2: M2 macrophages; TNF: tumor necrosis factor; HCC: hepatocellular carcinoma; IFN: interferon; IL: interleukin; SPNS2: spinster homologue 2; TCGA: The Cancer Genome Atlas. |
SPNS2 expression in HCC tumors was associated with enrichment of tumor-aggravating pathways, but with less cell proliferation
As we had previously found that SPNS2 expression correlated with lymphangiogenesis, we then conducted GSEA to evaluate the link between SPNS2 expression and other tumor-aggravating pathways. We found that many of the cancer-aggravating pathways were enriched with high SPNS2 expression, such as KRAS signaling, epithelial-mesenchymal transition (EMT), Notch signaling, tumor growth factor (TGF)-β signaling, apical junction, hypoxia, androgen response, and estrogen response, consistently across both cohorts (FDR < 0.25 for all) (Fig. 4a).
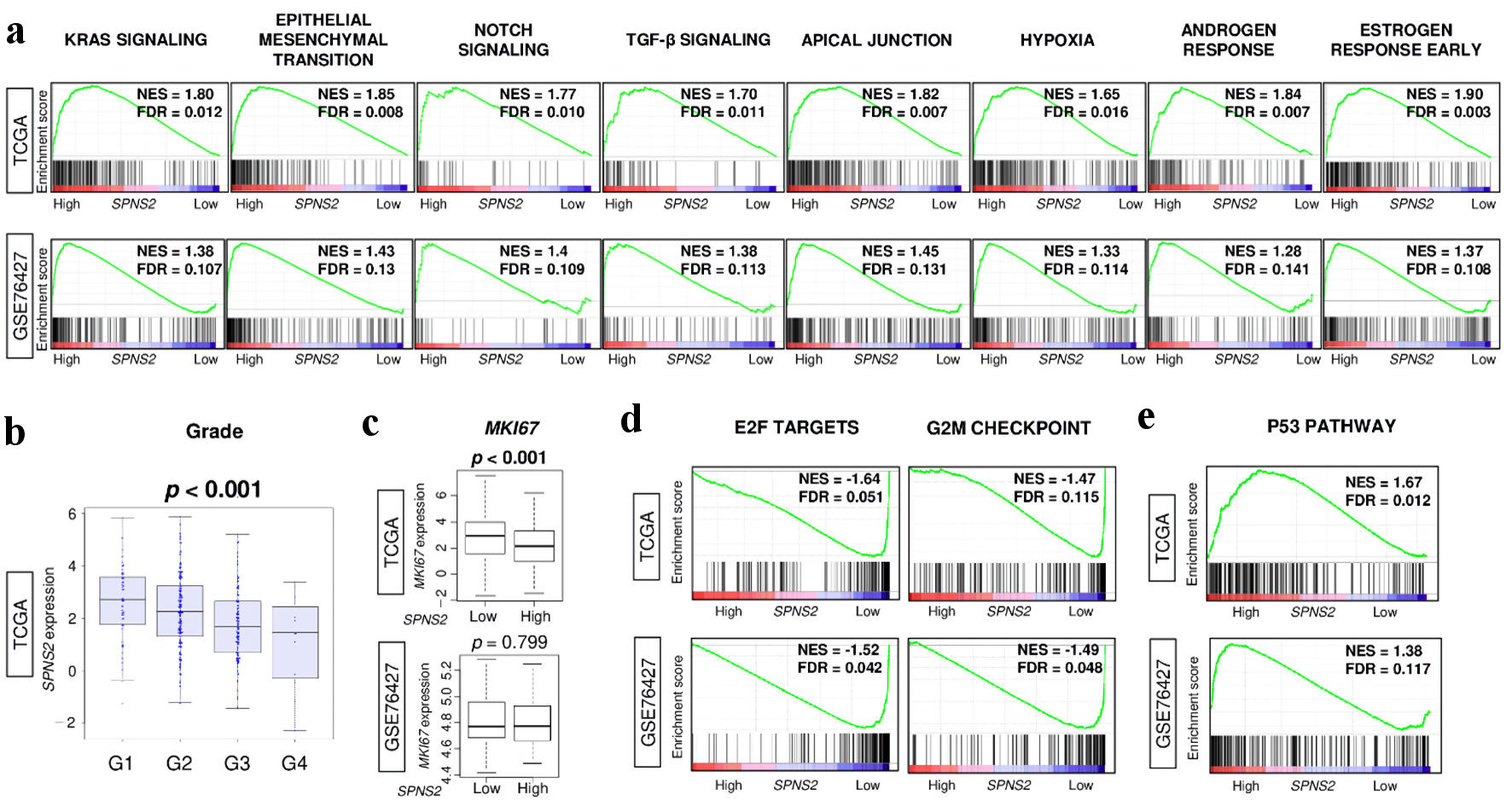 Click for large image | Figure 4. Tumor aggravating pathways were enriched to high SPNS2-expressing HCC, but it was associated with less cell proliferation. (a) Gene set enrichment analysis (GSEA) of tumor aggravating pathways in hallmark collection; KRAS signaling, epithelial mesenchymal transition (EMT), Notch signaling, TGF-β signaling, apical junction, hypoxia, androgen response, and estrogen response early. (b) SPNS2 gene expression levels by Nottingham histological grade in TCGA cohort. (c) Ki67 gene (MKI67) expression levels by high vs. low expression of SPNS2. GSEA of (d) cell proliferation-related gene sets in hallmark collection; E2F targets and G2M checkpoint, and (e) p53 pathway. FDR of less than 0.25 were defined as statistically significant following the recommendation by the Broad Institute. NES: normalized enrichment score; FDR: false discovery rate; E2F: cellular DNA binding activity regulating expression of E2 promoter; G2M: G2/M DNA damage checkpoint; HCC: hepatocellular carcinoma; KRAS: Kirsten rat sarcoma virus; MKI67: marker of proliferation Ki-67; SPNS2: spinster homologue 2; TCGA: The Cancer Genome Atlas; TGF: tumor growth factor. |
However, this did not translate to cancer cell proliferation. We found that SPNS2 expression actually negatively correlated with tumor grade in TCGA cohort (P < 0.001) (Fig. 4b). Additionally, HCCs with high SPNS2 expression had lower MKI67 expression (P < 0.001), indicating less cell proliferation (Fig. 4c). In order to investigate this finding further, we conducted GSEA for cell proliferation-related pathways and found that E2F targets and G2M checkpoint were enriched with lower SPNS2 expression, not higher, consistently in both cohorts (FDR < 0.25 for all) (Fig. 4d). Additionally, in conducting GSEA for the tumor suppressing p53 pathway, we found that HCCs with higher expression of SPNS2 enriched the p53 pathway in both cohorts (FDR < 0.25 for both) (Fig. 4e). Thus, although SPNS2 expression correlates with expression of multiple tumor-aggravating pathways, it is actually linked to lower tumor grade and proliferation.
The TME of SPNS2-high tumors is characterized by higher stromal cell infiltration
Given the finding that SPNS2 expression was associated with less cell proliferation, we aimed to gain a broader view of the TME by comparing the infiltration of stromal cells in SPNS2-high versus SPNS2-low HCCs using the xCell algorithm. We found that increased SPNS2 expression correlated with a higher infiltration of fibroblasts and adipocytes in the TCGA cohort (P < 0.001), and with adipocyte infiltration in the GSE74627 cohort (P = 0.01) (Fig. 5a). Subsequently, we conducted GSEA of adipogenesis and fatty acid metabolism gene sets comparing high versus low SPNS2 expression and found that they were significantly enriched to SPNS2-high HCC consistently across both TCGA and GSE78427 cohorts (FDR < 0.25) (Fig. 5b). These results show that high SPNS2 expressing HCC patients were associated with increased generation of adipocytes and infiltration of fibroblasts in the TME, which is in agreement with less cancer cell proliferation.
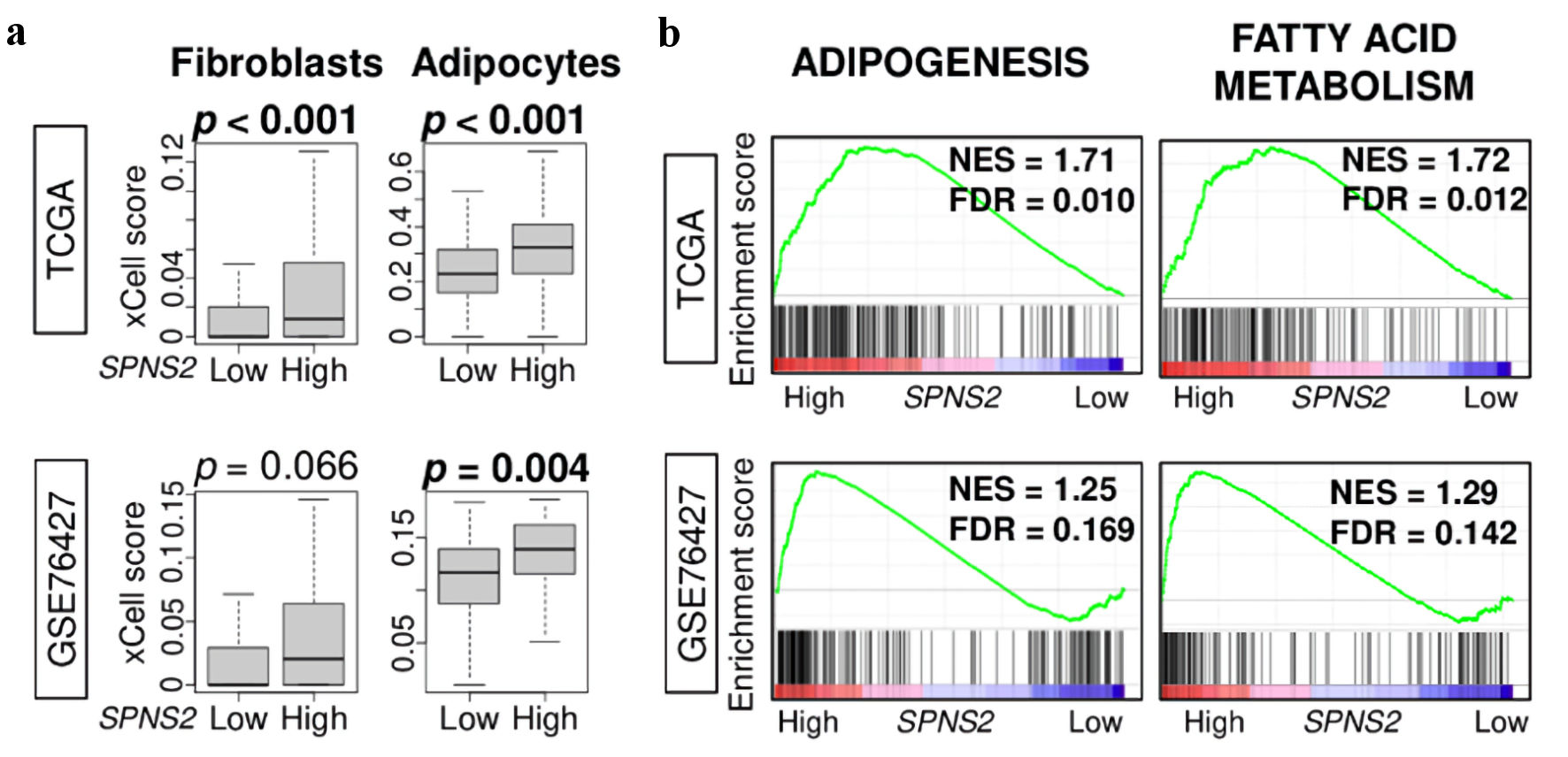 Click for large image | Figure 5. Stromal cell infiltration and metabolism related gene sets. (a) Estimated abundance of stromal cells using xCell algorithm. (b) Gene set enrichment analysis (GSEA) of metabolism-related pathways: adipogenesis, and fatty acid metabolism. The left side represents high expression, and the right side represents low expression of SPNS2. SPNS2 high and low expression groups were divided by median to perform the analysis. Normalized enrichment score (NES) and false discovery rate (FDR) of each analysis are shown in the graph. FDR of less than 0.25 were defined as statistically significant following the recommendation of the developer. SPNS2: spinster homologue 2; TCGA: The Cancer Genome Atlas. |
High SPNS2 expression was associated with better disease-free survival (DFS) in HCC patients of TCGA cohort
We initially hypothesized that HCC with high expression of SPNS2 would lead to worse survival. Surprisingly, we found that high expression of SPNS2 was associated with significantly better DFS (P = 0.031), disease-specific survival (DSS) (P = 0.046) and overall survival (OS) (P = 0.040) in the TCGA cohort (Fig. 6). This result was validated by Kaplan-Meier Plotter (RNA-seq, n = 364 [43]) that patients with high SPNS2 expression HCC had better OS, hazard ratio (HR) = 0.52 (0.36 - 0.75), log-rank P = 0.00033. On the other hand, although DFS was significantly better in high SPNS2 with log-rank P = 0.042, HR (high) = 0.73, P (HR) = 0.041, n (high) = 182 and n (low) = 182, there was no difference in OS (log-rank P = 0.082, HR (high) = 0.73, P (HR) = 0.083, n (high) = 182 and n (low) = 182) in Gene Expression Profiling Interactive Analysis (GEPIA) [44].
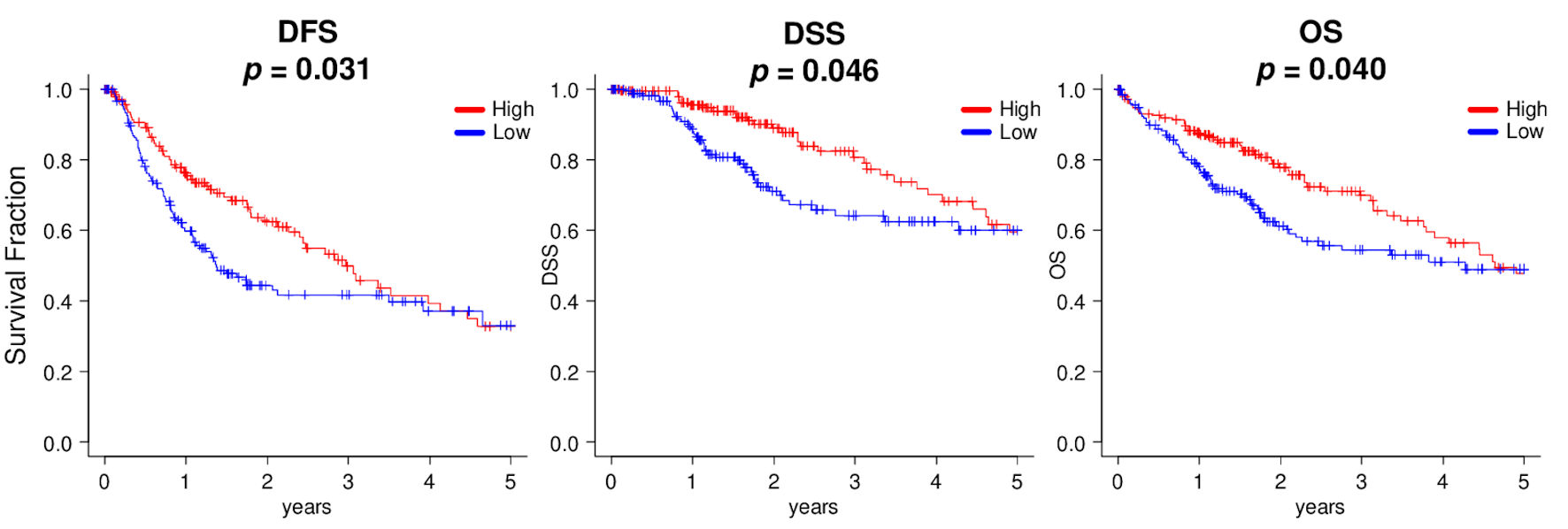 Click for large image | Figure 6. Kaplan-Meier survival analyses between high (red line) and low (blue line) expressions of SPNS2, divided by median cutoff in HCC patients of TCGA. DFS: disease-free survival; DSS: disease-specific survival; OS: overall survival. |
| Discussion | ▴Top |
S1P has been linked to inflammation-related cancers such as HCC, and this association has been explored in our previous works [8] as well as extensively by several other groups. Since S1P is a signaling lipid, many studies measured the expression of specific generating kinases, sphingosine kinase 1 and 2. In the current study, we focused on SPNS2 because it is a specific S1P transporter [16], and we have previously shown that it is associated with lymphangiogenesis [45] that plays a role in cancer progression. SPNS2 plays a complex role in cancer progression both linked to and independent of its effect on S1P concentrations in the TME.
In our study, we found that SPNS2 expression correlated with lymphatic endothelial cell infiltration as well as increased lymphangiogenesis and angiogenesis. Gene expressions of CXCR4 and PROX1 showed a similar trend but did not reach significance in validation cohort, most likely due to its small cohort size. This aligns with previous studies which have demonstrated that SPNS2-deficient mice have irregularity and disorganization of lymphatic vessels [45, 46], suggesting that SPNS2 is required for lymphatic vessel maturation. Given the enrichment of angiogenesis gene set without infiltrations of microvascular endothelial cells but with pericytes, this implicates enhanced generation of mature blood vessels in SPNS2-high HCCs. As we identified a link between SPNS2 expression and lymphangiogenesis/angiogenesis, we evaluated the effect of SPNS2 expression on other tumor-promoting pathways. Indeed, we found as expected that many pro-cancer pathways such as KRAS signaling and EMT were enriched in high-SPNS2 HCC tumors. Interestingly, our findings here are in contrast with a 2021 study which found that SPNS2 expression in colorectal cancer (CRC) was negatively correlated with EMT [47]. This suggests that the effect of SPNS2 on tumor progression could vary depending on the type of cancer.
Based on our findings that SPNS2 expression correlated with expression of cancer-promoting pathways, we expected to find that tumor grade, size and proliferation would be greater in high-SPNS2 HCC. Surprisingly, we found that the converse was true. Although SPNS2 expression trended upward in correlation with progressive liver tissue dysplasia and fibrosis, it then peaked in early HCC and was then inversely correlated with tumor grade and size. SPNS2 also inversely correlated with MKI67 expression and enrichment of cell proliferation pathways. Our findings suggest that SPNS2 may play different roles at various stages of tumor development, with initial stimulation of lymphangiogenesis that increases with worsening dysplasia and inflammation, but afterward with tumor suppression effects in higher stage disease. Lv et al also found that SPNS2 expression was significantly lower in higher stage CRC tumors and in liver metastases from CRC, concluding that the eventual downregulation of SPNS2 is important to the progression of CRC [47].
In addition to its role in aggravating cancer, S1P and its transporter SPNS2 have been well described in regard to their roles in immune system function and inflammation [15, 16]. For example, mice with SPNS2 gene deletion have been shown to have reduced inflammation [16], and S1P has been linked to TNF-α release [48] and fibrosis [49, 50]. SPNS2 expression is known to play a role in B- and T-lymphocyte egress [5, 16, 51-53], as well as in the survival of naive T cells [54]. Additionally, SPNS2 expression and the S1P gradient it generates has been found to be essential to the appropriate positioning of natural killer (NK) cells within the periphery of lymph nodes, which in turn is important for timely macrophage stimulation via IFN-γ [16, 55]. Lastly, as lymphatic endothelial cell expression of SPNS2 is specific to secretion of lymph S1P without effect on concentrations of plasma S1P, SPNS2 has been suggested as a potential target of immunotherapy [56, 57]. Surprisingly, we found that high SPNS2 HCC was not associated with significantly high infiltration of specific immune cell infiltration in the TME as would be expected. While SPNS2-high tumors showed lower cell scores of anti-cancer immune cells CD8, Th1, and M1, they also demonstrated lower cell scores of pro-cancer immune cells Th2 and B cells. These results demonstrate that, although higher levels of S1P export were significantly associated with host immune response, there was no net influence of SPNS2 expression on the immune cell infiltration, suggesting the involvement of additional factors. For example, CD4+ CD25+ regulatory T cells (Tregs) are known to dampen the immune response by suppressing self-reactive T lymphocytes and have a well-described role in HCC in part due to the liver’s function as a central immunomodulator [58]. However, the role of Tregs in HCC varies depending on the underlying etiology; in chronic viral hepatitis, an increase in peripheral and liver Tregs have been reported, whereas autoimmune liver disease is characterized by a qualitative and quantitative Treg deficiency [58]. Further, infiltration of Tregs may even parallel other T cells in certain occasions [59]. Given that our cohorts included a variety of liver tissue samples with various underlying pathologies, this may partly explain the inconsistency of immune cell infiltration in the TME.
On the other hand, high-SPNS2 HCCs consistently showed significant enrichment of inflammatory and immune-response related gene sets. We did find that high-SPNS2 tumors showed a higher infiltration of adipocytes and fibroblasts, which is in line with prior studies demonstrating that the function of SPNS2 in stromal cells is crucial to immune system function [60]. Although it is possible that chronic inflammation that generated HCC may have evoked lymphangiogenesis via SPNS2 expression, this causal relationship needs to be proved by experimental settings.
Finally, our results demonstrated that higher expression of SPNS2 was unexpectedly linked to a statistically significant increase in DFS, and a trend toward better DSS and OS. This was surprising to us given the studies which have demonstrated that deficiency of the SPNS2 gene is associated with metastatic suppression [57, 61]. These results were validated by Kaplan-Meier Plotter analysis and GEPIA, although OS was not statistically significant, which may be due to other cohort factors such as age or comorbidities of the patients. As noted above, these findings may reflect the different roles that SPNS2 may play at various stages of tumor development, and this highlights the complexity of the interactions within the TME. Taken together, we cannot help but speculate that high SPNS2-expressing HCC that secrete S1P evoke immune response without specific infiltration of immune cells, which may have had a positive impact on response to treatment or suppression of recurrence of HCC that overcame the detrimental effects of tumor aggravating pathways.
The link between SPNS2 and lymphangiogenesis as well as immune-response related gene sets suggests its potential as a target in treatment of HCC. Historically, systemic treatment of HCC was limited to the tyrosine kinase inhibitor (TKI) sorafenib; and despite the impact that immune checkpoint inhibitor (ICI) therapy has had on other cancers such as melanoma and lung cancer, ICI monotherapy was not found to provide a survival benefit in HCC compared to sorafenib [4]. However, the recent IMbrave150 trial demonstrated superiority of combined ICI atezolizumab plus anti-vascular epithelial growth factor (VEGF) monoclonal antibody bevacizumab in the treatment of unresectable HCC, which has stimulated interest in combination therapy of TKIs plus ICIs for HCC, with several ongoing clinical trials [4]. The success of combination therapy in HCC plays on the interaction between proangiogenic factors and the anticancer immune response, both of which are shown in our results to be associated with SPNS2 expression.
There are some limitations associated with the use of a transcriptomic database. The retrospective nature of the study and the narrow clinical picture obtained from a database do restrict our ability to define the causal association between SPNS2 expression and tumor progression. Many factors can potentially affect survival. For example, a 2021 epidemiological study out of Italy spanning the previous 15 years demonstrated the changing scenario of HCC in the last decades, showing that patient survival improved over time due to increased use of thermal ablation as well as patient-tailored therapy in intermediate stages [62]. Unfortunately, these factors cannot be easily captured in a tissue sample database. On the other hand, the use of a transcriptomic database and a computational algorithm for examining cell composition afforded us a larger sample size as well as the ability to obtain a global picture of the TME.
In conclusion, we found that while increased expression of the SPNS2 transporter in HCC was associated with lymphangiogenesis and multiple tumor-promoting cancer pathways, SPNS2 expression clinically correlated with decreased cancer cell proliferation and an improved DFS. Our findings help to further illustrate the relationship between the S1P axis and tumor progression in HCC.
Acknowledgments
None to declare.
Financial Disclosure
KT was supported by the US NIH grant R01CA160688. Roswell Park Comprehensive Cancer Center is supported by NCI/NIH grant P30-CA016056.
Conflict of Interest
The authors have no potential conflict of interest to disclose.
Informed Consent
Not applicable.
Author Contributions
JS: writing - original draft. MO: writing - original draft, data collection, data curation, and formal analysis. VS: writing - original draft. KC: data curation, and formal analysis. LY: formal analysis, and supervision (bioinformatics). AM: interpretation of analysis (SPNS2). NH: supervision (interpretation of data on SPNS2). IE: supervision (interpretation of data on HCC). KT: conceptualization, visualization, and writing - review and editing.
Data Availability
The data supporting the findings of this study are publicly available in TCGA and GEO and can be accessed via https://portal.gdc.cancer.gov/.
Abbreviations
ANOVA: analysis of variance; CAS: cytolytic activity score; CRC: colorectal cancer; CXCL: chemokine (C-X-C motif) ligand; CXCR: chemokine (C-X-C motif) receptor; DFS: disease-free survival; DSS: disease-specific survival; E2F: cellular DNA binding activity regulating expression of E2 promoter; EMT: epithelial-mesenchymal transition; FDR: false discovery rate; G2M: G2/M DNA damage checkpoint; GEO: Gene Expression Omnibus; GSE: gene set enrichment; GSEA: gene set enrichment analysis; HCC: hepatocellular carcinoma; IFN: interferon; IL: interleukin; JAK/STAT: Janus kinase/signal transducer and activator of transcription; KRAS: Kirsten rat sarcoma virus; LYVE: lymphatic vessel endothelial hyaluronic acid receptor; MKI67: marker of proliferation Ki-67; MSigDB: Molecular Signatures Database; NK: natural killer; OS: overall survival; PECAM: platelet endothelial cell adhesion molecule; PROX: Prospero homeobox; S1P: sphingosine-1-phosphate; SOX: Sry-type HMG box; SPNS2: spinster homologue 2; TCGA: The Cancer Genome Atlas; TGF: tumor growth factor; TME: tumor microenvironment; TNF: tumor necrosis factor
| References | ▴Top |
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7-33.
doi pubmed - Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1-61.
doi pubmed pmc - Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: From diagnosis to treatment. Surg Oncol. 2016;25(2):74-85.
doi pubmed - Stefanini B, Ielasi L, Chen R, Abbati C, Tonnini M, Tovoli F, Granito A. TKIs in combination with immunotherapy for hepatocellular carcinoma. Expert Rev Anticancer Ther. 2023;23(3):279-291.
doi pubmed - Spiegel S, Maczis MA, Maceyka M, Milstien S. New insights into functions of the sphingosine-1-phosphate transporter SPNS2. J Lipid Res. 2019;60(3):484-489.
doi pubmed pmc - Takabe K, Paugh SW, Milstien S, Spiegel S. "Inside-out" signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol Rev. 2008;60(2):181-195.
doi pubmed pmc - Tsuchida J, Nagahashi M, Nakajima M, Moro K, Tatsuda K, Ramanathan R, Takabe K, et al. Breast cancer sphingosine-1-phosphate is associated with phospho-sphingosine kinase 1 and lymphatic metastasis. J Surg Res. 2016;205(1):85-94.
doi pubmed pmc - Nagahashi M, Abe M, Sakimura K, Takabe K, Wakai T. The role of sphingosine-1-phosphate in inflammation and cancer progression. Cancer Sci. 2018;109(12):3671-3678.
doi pubmed pmc - Takabe K, Spiegel S. Export of sphingosine-1-phosphate and cancer progression. J Lipid Res. 2014;55(9):1839-1846.
doi pubmed pmc - Yamada A, Nagahashi M, Aoyagi T, Huang WC, Lima S, Hait NC, Maiti A, et al. ABCC1-Exported Sphingosine-1-phosphate, Produced by Sphingosine Kinase 1, shortens survival of mice and patients with breast cancer. Mol Cancer Res. 2018;16(6):1059-1070.
doi pubmed pmc - Maiti A, Takabe K, Hait NC. Metastatic triple-negative breast cancer is dependent on SphKs/S1P signaling for growth and survival. Cell Signal. 2017;32:85-92.
doi pubmed pmc - Xu G, Yang Z, Sun Y, Dong H, Ma J. Interaction of microRNAs with sphingosine kinases, sphingosine-1 phosphate, and sphingosine-1 phosphate receptors in cancer. Discov Oncol. 2021;12(1):33.
doi pubmed pmc - Aoyagi T, Nagahashi M, Yamada A, Takabe K. The role of sphingosine-1-phosphate in breast cancer tumor-induced lymphangiogenesis. Lymphat Res Biol. 2012;10(3):97-106.
doi pubmed pmc - Huang WC, Nagahashi M, Terracina KP, Takabe K. Emerging Role of Sphingosine-1-phosphate in Inflammation, Cancer, and Lymphangiogenesis. Biomolecules. 2013;3(3):408-434.
doi pubmed pmc - Hisano Y, Nishi T, Kawahara A. The functional roles of S1P in immunity. J Biochem. 2012;152(4):305-311.
doi pubmed - Aoki M, Aoki H, Ramanathan R, Hait NC, Takabe K. Sphingosine-1-phosphate signaling in immune cells and inflammation: roles and therapeutic potential. Mediators Inflamm. 2016;2016:8606878.
doi pubmed pmc - Kawahara A, Nishi T, Hisano Y, Fukui H, Yamaguchi A, Mochizuki N. The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science. 2009;323(5913):524-527.
doi pubmed - Donoviel MS, Hait NC, Ramachandran S, Maceyka M, Takabe K, Milstien S, Oravecz T, et al. Spinster 2, a sphingosine-1-phosphate transporter, plays a critical role in inflammatory and autoimmune diseases. FASEB J. 2015;29(12):5018-5028.
doi pubmed pmc - Li M, Tang Y, Wang D, Zhai X, Shen H, Zhong C, Yao M, et al. Sphingosine-1-phosphate transporter spinster homolog 2 is essential for iron-regulated metastasis of hepatocellular carcinoma. Mol Ther. 2022;30(2):703-713.
doi pubmed pmc - Fang L, Hou J, Cao Y, Shan JJ, Zhao J. Spinster homolog 2 in cancers, its functions and mechanisms. Cell Signal. 2021;77:109821.
doi pubmed - Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400-416.e411.
doi pubmed pmc - Gandhi S, Elkhanany A, Oshi M, Dai T, Opyrchal M, Mohammadpour H, Repasky EA, et al. Contribution of immune cells to glucocorticoid receptor expression in breast cancer. Int J Mol Sci. 2020;21(13):4635.
doi pubmed pmc - Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, et al. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in hepatocellular carcinoma (HCC). Cancers (Basel). 2021;13(2):323.
doi pubmed pmc - Takahashi H, Katsuta E, Yan L, Tokumaru Y, Katz MHG, Takabe K. Transcriptomic profile of lymphovascular invasion, a known risk factor of pancreatic ductal adenocarcinoma metastasis. Cancers (Basel). 2020;12(8):2033.
doi pubmed pmc - Grinchuk OV, Yenamandra SP, Iyer R, Singh M, Lee HK, Lim KH, Chow PK, et al. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol Oncol. 2018;12(1):89-113.
doi pubmed pmc - Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938-947.
doi pubmed - http://software.broadinstitute.org/gsea/index.jsp.
- Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, et al. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9(7):1643.
doi pubmed pmc - Schulze A, Oshi M, Endo I, Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int J Mol Sci. 2020;21(21):8127.
doi pubmed pmc - Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, et al. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10(1):1852.
doi pubmed pmc - Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, et al. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10(3):897-907.
pubmed pmc - Katsuta E, Rashid OM, Takabe K. Fibroblasts as a biological marker for curative resection in pancreatic ductal adenocarcinoma. Int J Mol Sci. 2020;21(11):3890.
doi pubmed pmc - Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, et al. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21(18):6968.
doi pubmed pmc - Tokumaru Y, Asaoka M, Oshi M, Katsuta E, Yan L, Narayanan S, Sugito N, et al. High expression of microRNA-143 is associated with favorable tumor immune microenvironment and better survival in estrogen receptor positive breast cancer. Int J Mol Sci. 2020;21(9):3213.
doi pubmed pmc - Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
doi pubmed pmc - Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-457.
doi pubmed pmc - Wu R, Sarkar J, Tokumaru Y, Takabe Y, Oshi M, Asaoka M, Yan L, et al. Intratumoral lymphatic endothelial cell infiltration reflecting lymphangiogenesis is counterbalanced by immune responses and better cancer biology in the breast cancer tumor microenvironment. Am J Cancer Res. 2022;12(2):504-520.
pubmed pmc - Oshi M, Huyser MR, Le L, Tokumaru Y, Yan L, Matsuyama R, Endo I, et al. Abundance of microvascular endothelial cells is associated with response to chemotherapy and prognosis in colorectal cancer. Cancers (Basel). 2021;13(6):1477.
doi pubmed pmc - Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, et al. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21(18):6708.
doi pubmed pmc - Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, et al. Plasmacytoid dendritic cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in triple negative breast cancer (TNBC) more strongly than conventional dendritic cell (cDC). Cancers (Basel). 2020;12(11):3342.
doi pubmed pmc - Wu R, Oshi M, Asaoka M, Yan L, Benesch MGK, Khoury T, Nagahashi M, et al. Intratumoral Tumor Infiltrating Lymphocytes (TILs) are associated with cell proliferation and better survival but not always with chemotherapy response in breast cancer. Ann Surg. 2023;278(4):587-597.
doi pubmed pmc - http:///www.r-project.org/.
- https://kmplot.com/analysis/index.php?p=service.
- http://gepia.cancer-pku.cn/detail.php?gene=&clicktag=survival###.
- Nagahashi M, Kim EY, Yamada A, Ramachandran S, Allegood JC, Hait NC, Maceyka M, et al. Spns2, a transporter of phosphorylated sphingoid bases, regulates their blood and lymph levels, and the lymphatic network. FASEB J. 2013;27(3):1001-1011.
doi pubmed pmc - Pham TH, Baluk P, Xu Y, Grigorova I, Bankovich AJ, Pappu R, Coughlin SR, et al. Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J Exp Med. 2010;207(1):17-27.
doi pubmed pmc - Lv L, Yi Q, Yan Y, Chao F, Li M. SPNS2 downregulation induces EMT and promotes colorectal cancer metastasis via activating AKT signaling pathway. Front Oncol. 2021;11:682773.
doi pubmed pmc - Terlizzi M, Colarusso C, Ferraro G, Monti MC, Cerqua I, Roviezzo F, Pinto A, et al. Sphingosine-1-phosphate contributes to TLR9-induced TNF-alpha release in lung tumor cells. Cell Physiol Biochem. 2021;55(2):222-234.
doi pubmed - Bernacchioni C, Ciarmela P, Vannuzzi V, Greco S, Vannuccini S, Malentacchi F, Pellegrino P, et al. Sphingosine 1-phosphate signaling in uterine fibroids: implication in activin A pro-fibrotic effect. Fertil Steril. 2021;115(6):1576-1585.
doi pubmed - Tanaka S, Zheng S, Kharel Y, Fritzemeier RG, Huang T, Foster D, Poudel N, et al. Sphingosine 1-phosphate signaling in perivascular cells enhances inflammation and fibrosis in the kidney. Sci Transl Med. 2022;14(658):eabj2681.
doi pubmed pmc - Okuniewska M, Fang V, Baeyens A, Raghavan V, Lee JY, Littman DR, Schwab SR. SPNS2 enables T cell egress from lymph nodes during an immune response. Cell Rep. 2021;36(2):109368.
doi pubmed pmc - Fukuhara S, Simmons S, Kawamura S, Inoue A, Orba Y, Tokudome T, Sunden Y, et al. The sphingosine-1-phosphate transporter Spns2 expressed on endothelial cells regulates lymphocyte trafficking in mice. J Clin Invest. 2012;122(4):1416-1426.
doi pubmed pmc - Hisano Y, Kobayashi N, Yamaguchi A, Nishi T. Mouse SPNS2 functions as a sphingosine-1-phosphate transporter in vascular endothelial cells. PLoS One. 2012;7(6):e38941.
doi pubmed pmc - Mendoza A, Fang V, Chen C, Serasinghe M, Verma A, Muller J, Chaluvadi VS, et al. Lymphatic endothelial S1P promotes mitochondrial function and survival in naive T cells. Nature. 2017;546(7656):158-161.
doi pubmed pmc - Fang V, Chaluvadi VS, Ramos-Perez WD, Mendoza A, Baeyens A, Rivera R, Chun J, et al. Gradients of the signaling lipid S1P in lymph nodes position natural killer cells and regulate their interferon-gamma response. Nat Immunol. 2017;18(1):15-25.
doi pubmed pmc - Mendoza A, Breart B, Ramos-Perez WD, Pitt LA, Gobert M, Sunkara M, Lafaille JJ, et al. The transporter Spns2 is required for secretion of lymph but not plasma sphingosine-1-phosphate. Cell Rep. 2012;2(5):1104-1110.
doi pubmed pmc - van der Weyden L, Arends MJ, Campbell AD, Bald T, Wardle-Jones H, Griggs N, Velasco-Herrera MD, et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature. 2017;541(7636):233-236.
doi pubmed pmc - Granito A, Muratori L, Lalanne C, Quarneti C, Ferri S, Guidi M, Lenzi M, et al. Hepatocellular carcinoma in viral and autoimmune liver diseases: Role of CD4+ CD25+ Foxp3+ regulatory T cells in the immune microenvironment. World J Gastroenterol. 2021;27(22):2994-3009.
doi pubmed pmc - Oshi M, Sarkar J, Wu R, Tokumaru Y, Yan L, Nakagawa K, Ishibe A, et al. Intratumoral density of regulatory T cells is a predictor of host immune response and chemotherapy response in colorectal cancer. Am J Cancer Res. 2022;12(2):490-503.
pubmed pmc - Nijnik A, Clare S, Hale C, Chen J, Raisen C, Mottram L, Lucas M, et al. The role of sphingosine-1-phosphate transporter Spns2 in immune system function. J Immunol. 2012;189(1):102-111.
doi pubmed pmc - Celia-Terrassa T, Kang Y. Mouse genomic screen reveals novel host regulator of metastasis. Genome Biol. 2017;18(1):31.
doi pubmed pmc - Garuti F, Neri A, Avanzato F, Gramenzi A, Rampoldi D, Rucci P, Farinati F, et al. The changing scenario of hepatocellular carcinoma in Italy: an update. Liver Int. 2021;41(3):585-597.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


