| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 1, February 2024, pages 100-113
Inhibiting the m6A Reader IGF2BP3 Suppresses Ovarian Cancer Cell Growth via Regulating PLAGL2 mRNA Stabilization
Tian Tian Daia, f, Yi Ze Lib, f, Hui Ting Huc, f, Yong Mei Zhaod, Hong Yan Penge, Wen Dong Baid, g, Jing Wen Wanga, g
aDepartment of Pharmacy, Xijing Hospital, Fourth Military Medical University, Xi’an 710032, Shaanxi, China
bDepartment of Clinical Oncology, Xijing Hospital, Fourth Military Medical University, Xi’an 710032, Shaanxi, China
cSchool of Medicine and Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing 210023, Jiangsu, China
dDepartment of Hematology, Xinjiang Command General Hospital of Chinese People’s Liberation Army, Urumqi 830000, Xinjiang, China
eDepartment of Internal Medicine, 63650 Military Hospital, Urumqi 830000, Xinjiang, China
fThese authors contributed equally to the study.
gCorresponding Author: Jing Wen Wang, Department of Pharmacy, Xijing Hospital, Fourth Military Medical University, Xi’an 710032, Shaanxi, China; Wen Dong Bai, Department of Hematology, Xinjiang Command General Hospital of Chinese People’s Liberation Army, Urumqi 830000, Xinjiang, China
Manuscript submitted October 22, 2023, accepted December 1, 2023, published online January 10, 2024
Short title: IGF2BP3 Promotes OC via m6A Modification of PLAGL2
doi: https://doi.org/10.14740/wjon1747
| Abstract | ▴Top |
Background: The oncogene IGF2 mRNA binding protein 3 (IGF2BP3) could function as an m6A reader in stabilizing many tumor-associated genes’ mRNAs. However, the relevant oncogenic mechanism by which IGF2BP3 promotes ovarian cancer growth is largely unknown.
Methods: The IGF2BP3 expression in ovarian cancer was identified by retrieving the datasets from The Cancer Genome Atlas (TCGA). GEO datasets evaluated the relevant signaling pathways in IGF2BP3 knockdown in ovarian cancer cells. IGF2BP3 positive correlation gene in TCGA was calculated. MTS proliferation assay was identified in IGF2BP3 knockdown and rescued by PLAG1 like zinc finger 2 (PLAGL2) overexpression in ES-2 and SKOV3 cells. Bioinformatic analysis and RIP-qPCR were predicted and identified the IGF2BP3 binding site and PLAGL2 mRNA stability. The animal experiment identified IGF2BP3 proliferation inhibition.
Results: IGF2BP3 was upregulated in ovarian cancer tissue and cells. The depletion of IGF2BP3 in ovarian cancer cells leads to an enhancement of the pathway involved in cellular proliferation and mRNA stability. IGF2BP3 positive correlation suppressed pro-proliferation gene PLAGL2. IGF2BP3 knockdown suppressed ovarian cancer cell proliferation and was rescued by PLAGL2 overexpression. Luciferase reporter assay confirmed that IGF2BP3 could bind to 3'-UTR of PLAGL2 to maintain the mRNA stability. Further, in in vivo experiments, IGF2BP3 knockdown suppressed ovarian cancer cell proliferation via inhibiting PLAGL2 expression.
Conclusion: All of these indicate that PLAGL2 mediates the main function of IGF2BP3 knockdown on ovarian cancer proliferation inhibition through mRNA stability regulation.
Keywords: Ovarian cancer; IGF2BP3; mRNA stability; m6A methylation; PLAGL2
| Introduction | ▴Top |
N6-methyladenosine (m6A) is the predominant modification found in RNA and plays a crucial role in regulating diverse post-transcriptional processes, including mRNA stability, translocation, and protein translation efficiency. As a result, it exerts a significant influence on numerous biological processes, such as cell proliferation, tumorigenesis, and interactions between RNA and proteins [1]. These processes involve the participation of m6A methyltransferases (“writers”), demethylases (“erasers”), and RNA-binding proteins (“readers”) in dynamic regulation of m6A modifications. The insulin-like growth factor 2 mRNA-binding protein family (IGF2BPs), among other readers, are capable of recognizing m6A-mediated physiological behavior and exerting an influence on RNA function [2]. Meanwhile, methyltransferases, such as methyltransferase like protein 3 (METTL3) or METTL14, can complex various scaffold proteins to form writers to regulate RNA methylation [3]. Aside from this, ovarian cancer (OC) is the leading cause of death from all gynecologic malignancies [4]. Recently, abnormally expressed m6A regulators have been linked to OC initiation and progression [3]. Therefore, understanding the role of m6A modification in ovarian tumorigenesis is urgent for diagnosis and therapy development.
The RNA-binding protein, IGF2BP3, plays a role in the regulation of mRNA stability during carcinogenesis [1]. There is, however, little information about their paralogue-specific functions in tumor cells [5]. IGF2BP3 has recently been identified as a reader of m6A modification, exhibiting a preference for binding to N6-methyladenosine-modified target mRNAs [1]. Meanwhile, IGF2BPs were shown to regulate the mRNA stability and modulate neural stem cell proliferation [6]. However, the biological role of IGF2BP3 during OC development remains unclear.
In the present study, online databases profile was analyzed and found an up-regulation of IGF2BP3 associated with poor progression-free survival (PFS). Further mechanistic study demonstrated that IGF2BP3 regulates OC cell proliferation via modulating the mRNA stability of the downstream gene PLAGL2.
| Materials and Methods | ▴Top |
Specimens and cell culture
The cancerous and normal ovarian specimens were procured from patients who received treatment at the Department of Oncology of Xijing Hospital during the period from March to December 2022. All participants included in the study were histologically confirmed to have serous ovarian cancer (n = 14), mucinous ovarian cancer (n = 1), endometrioid ovarian cancer (n = 3), and clear cell ovarian cancer (n = 2). Tissue samples were promptly obtained following resection and subsequently divided into three segments to facilitate RNA isolation, protein extraction, and storage in liquid nitrogen for subsequent analyses. Cells used in this research were obtained from the American Type Culture Collection (ATCC, USA). The study protocol received ethical approval from the Fourth Military Medical University affiliated Xijing Hospital’s ethics committee (registration number: KY20173012-1). The ES-2 and SKOV3 ovarian cancer cells were grown at 37 °C in incubators with 5% CO2 in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 10% fetal bovine serum.
Bioinformatics analysis
In order to measure the expression levels of IGF2BP3 and PLAGL2, the RNA-sequence transcriptome data and relative clinicopathological characteristics for 419 OC tissues and 88 adjacent tissues were obtained from the TCGA database[7]. The datasets for analyzing the effects of IGF2BP3 knockdown in OC cells on expression profiling were obtained from the GEO database (GSE109604). Data of IGF2BP3 expression in ovarian cancer tissues were obtained from the GEO database GSE154600 (single-cell sequencing) and GSE14407 (microarray). The Human Protein Atlas database was used to verify the protein level of IGF2BP3 [8]. Exploring the PLAGL2 enrichment data in m6A modification profiling sequencing of OC tissues was obtained from the GEO dataset (GSE119168). Enrichment bio-pathways were characterized using the ClueGO (version 2.4.1) and CluePedia (version 1.4.2) plugins in Cytoscape [3]. IGF2BP3 binding motifs were obtained from the RBPmap website [9]. IGF2BP3 m6A RNA methylation site on PLAGL2 3'-UTR was predicted in the online tool m6AVar website [10]. Kaplan-Meier survival curves were drawn in the online tool KMplot program website [11].
Immunohistochemistry (IHC)
IHC was performed following the previously described protocol [12]. Paraformaldehyde was used to fix the tissue samples, followed by embedding them in paraffin. Antigen retrieval for IHC analysis was achieved through heat-induced treatment with ethylenediaminetetraacetic acid (EDTA) buffer. To inhibit endogenous peroxidase activity, 3% H2O2 was utilized. The sections were then blocked with 5% goat serum for a duration of 1 h, followed by incubation with antibodies targeting IGF2BP3 (ab179807, 1:200, Abcam, MA, USA) or PLAGL2 (ab139509, 1:200, Abcam). Following this, the sections were subjected to a 1-h incubation period with the secondary antibody (BA1054, 1:1,500, Boster, China). Afterwards, the sections underwent processing using an ABC HRP kit and a DAB substrate kit (Zhongshan Jinqiao Biotechnology Co., Ltd, China). Nuclei were counterstained with hematoxylin. Subsequently, the sections were washed with water to eliminate any excess substrate, dehydrated, and finally covered with a mounting medium. The evaluation of images was performed using the IHC profiler plugin in ImageJ (NIH, USA).
CellTiter 96 AQueous one-solution cell proliferation assay (MTS)
ES-2 and SKOV3 cells were initially plated at a density of 5,000 cells per well in a 96-well plate and maintained in a standard growth medium for the designated time periods. This was followed by an evaluation of cell proliferation using the CellTiter 96 aqueous one-solution cell proliferation assay kit from Promega (MTS assay), according to manufacturer’s instructions. In order to maintain uniformity, the MTS signal values for each cell line were adjusted to account for the time elapsed since the cells were sub-cultured, approximately 5 or 24 h prior [13].
Plasmid construction and lentivirus packaging
The coding sequences of IGF2BP3 and PLAGL2 were amplified from cDNA extracted from SKOV3 cells and were constructed into the pCDNA3.0 vector (Invitrogen). The PLAGL2 untranslated region (3'-UTR) containing wildtype or mutated m6A modification site was amplified from ES-2-derived cDNA, and the resulting products were engineered into the luciferase reporter pGL3-basic plasmid (Promega). All gene modifications were validated by sequencing. The shRNA sequences and scramble sequence were designed and synthesized, followed by their cloning into pLKO.1 (Addgene, Cambridge, MA, USA; Plasmid 30323). Subsequently, lentivirus packaging was carried out in 293T cells. The virus present in the culture medium was obtained by collecting the supernatant and utilized for infecting the OC cells. After a transduction period of 48 h, the cells were subjected to selection using 5 mg/L Blasticidin S (Sigma, St. Louis, MO, USA) in order to establish stable cell lines. Primers used for plasmid construction and lentivirus packaging are as follows: IGF2BP3 ORF (F: 5'-ATGAACAAACTGTATATCGGAAAC-3', R: 5'-TTACTTCCGTCTTGACTGAGGTG-3'), PLAGL2 ORF (F: 5'-CATGACCACATTTTTCACCAG-3', R: 5'-TACTGGAATGCTTGATGGAAAC-3'), PLAGL2 3'-UTR (F:5'-CATCTTCCATGACTGCATTTG-3', R: 5'-CTTCCTCCTTTGCTATCCACAC-3'), PLAGL2 3'-UTR mut (F: 5'-CTCATCCTTCTCTTGAACTGGA-3', R: 5'-TTCTTCCCCTCTCCAGATCAAGA-3'), shIGF2BP3-1 sequence (5'-GCCTCATTCTTATTTCAAGAT-3'), shIGF2BP3-2 sequence (5'-CGGTGAATGAACTTCAGAATT-3'), shMETTL3 (5'-GCCTTAACATTGCCCACTGAT-3'), shMETTL14(5'-CCATGTACTTACAAGCCGATA-3'), and scramble (as control) (5'-TTCTCCGAACGTGTCACGT-3').
5-ethynyl-2′-deoxyuridine (EdU) incorporation assay
ES-2 and SKOV3 cells, which had been transduced with lentivirus, were seeded at a density of 4 × 103 cells/well into 96-well plates prior to treatment. Following the designated treatment, the cells were subjected to a 30-min pulse with EdU (Click-iT EdU imaging kits, Invitrogen, Paris, France) at room temperature, while being shielded from light. Following fixation in 2% paraformaldehyde, the cells were detected for EdU according to the manufacturer’s protocol [14]. DAPI was then used to stain the nuclei of the OC cells. For each dish, six fields were randomly selected using an imaging system microscope (Evos FL Auto 2, Invitrogen). The cells were counted using ImageJ software.
Dual luciferase reporter assays
An analysis of the interaction between PLAGL2 3'-UTR and IGF2BP3 or METTL14 was performed using the dual luciferase reporter assay kit (Promega, USA) as previously outlined [3]. Briefly, ES-2 cells were co-transfected vectors encoding mutated or wildtype PLAGL2 3'-UTR and IGF2BP3 or METTL14 using lipofectamine 2000. Meanwhile, plasmids expressing Renilla luciferase were transfected into each group of cells to normalize the transfection. After transfection, the cells were solubilized in the lysis buffer provided by the kit 48 h later, and luciferase activities from each group were determined using a multi-plate reader. An equation was developed to normalize relative activity to the activity of Renilla luciferase.
Quantitative real-time PCR analysis (RT-qPCR)
The cells were extracted from TRIzol reagent with SuperScript II reverse transcriptase (Invitrogen, USA) prior to RNA extraction. A RT-qPCR analysis was conducted in OC cells to determine IGF2BP3 and PLAGL2 expression levels. For RT-qPCR analysis, the SYBR Premix Ex Taq II from Takara was employed. The gene expression levels being studied were normalized against the abundance of GAPDH mRNA [15]. Primers are as follows: IGF2BP3 (F: 5'-TATATCGGAAACCTCAGCGAGA-3'; R: 5'-GGACCGAGTGCTCAACTTCT-3'); PLAGL2 (F: 5'- GAGTCAAGTGAAGTGCCAATGT-3'; R: 5'-TGAGGGCAGCTATATGGTCTC-3'); METTL3 (F: 5'- TTGTCTCCAACCTTCCGTAGT-3'; R: 5'-CCAGATCAGAGAGGTGGTGTAG-3'); METTL14 (F: 5'-GAACACAGAGCTTAAATCCCCA-3'; R: 5'- TGTCAGCTAAACCTACATCCCTG-3'); GAPDH (F: 5'- GGAGCGAGATCCCTCCAAAAT-3'; R: 5'-GGCTGTTGTCATACTTCTCATGG-3').
PLAGL2-specific m6A RT-qPCR analysis
The Magna MeRIP™ m6A Kit (17-10499, MERCK, USA) was used to determine the m6A modifications on PLAGL2, as previously described [2]. In brief, a total of 100 µg of RNA was subjected to metal-ion-induced fragmentation and subsequent purification. The resulting RNA was then subjected to incubation with either mouse IgG (ab190475, Abcam) or the antibody against m6A (ab284130, Abcam), followed by incubation with magnetic beads (ab214286, Abcam) at a temperature of 4 °C overnight. The immobilized methylated RNAs were subsequently precipitated, eluted, and recovered using the RNeasy kit (Qiagen). The fragmented RNA was then utilized for the detection of PLAGL2-specific m6A modification through RT-PCR, employing the following primers: F: 5'-TTCTCCAAGTACATTATGACC-3'; R: 5'-AGGGGATGGGGAGGTGACGG-3'.
Western blotting analysis
Western blotting was conducted by using the following antibodies: rabbit anti-IGF2BP3 (ab179807, 1:500, Abcam), rabbit anti-PLAGL2 (ab139509, 3:1,000, Abcam), rabbit anti-METTL3 (ab195352, 1:500, Abcam), rabbit anti-METTL14 (ab309096, 1:400, Abcam), and rabbit anti-β-actin (ab8827, 1:1,000, Abcam). The β-actin (ab7817, 1:2,000, Abcam) was employed as the internal control. This image was captured using a gel imaging system (Odyssey, LI-COR Biosciences, USA).
Measurement of mRNA half-life
ES-2 cells were subjected to treatment with actinomycin D (ACD, 5 µg/mL, ab291108, Abcam) for the specified durations (0, 1, 2, 3, and 4 days). Subsequently, RNA extraction and RT-qPCR were performed to assess the mRNA expression of PLAGL2 at each time interval. The PLAGL2 mRNA levels on days 1, 2, 3, and 4 were standardized against the level observed on day 0, enabling the evaluation of its half-life.
RNA immunoprecipitations (RIP)
For the RIP experiment, a total of 2 × 107 ES-2 cell extracts were prepared on ice using RIP buffer (ab156034, Abcam). The extracts were then sonicated on ice and treated with DNase for 30 min. Immunoprecipitation was carried out by incubating protein A/G precleared nuclear lysates with IgG (ab172730, Abcam), and IGF2BP3 (ab313556, Abcam) antibodies overnight. Following washing with RIP buffer, RNA/antibody complexes were eluted using SDS according to the standard protocol [16]. The co-purified RNA was extracted and quantified using qRT-PCR with the specified primers: METTL3 F: 5'-CCCCAAGGCTTCAACCAGG-3'; R: 5'-TATCTCCTGGCACTCGCAAGA-3'; METTL14 F: 5'- ACTGACCTAAAATCAGTCT-3'; R: 5'-TCAAACTGAGTCTTTGGTGG-3'.
RNA pulldown assays
IGF2BP3 was labeled using the Pierce™ RNA 3' end desthiobiotinylation kit (Pierce, USA) according to the provided protocol. The ES-2 lysate was incubated with either purified biotinylated PLAGL2 or anti-PLAGL2 at a temperature of 4 °C for a duration of 1 h. Subsequently, streptavidin-agarose beads were added to the lysate in order to precipitate the RNA-protein complexes. The beads were then subjected to washing with a washing buffer and eluted using laemmli buffer. Finally, a western blot assay was performed to ascertain the protein that interacted with the RNA.
Human OC xenografts in immunodeficient mice
The animal studies were carried out in compliance with the protocol that underwent review and approval by the Animal Care and Use Committee of the Fourth Military Medical University, adhering to the guidelines established by the National Institutes of Health (eighth edition, NIH, 2001). During the experiment, eight nude mice (6 - 8 weeks old) were randomly divided into two groups, and each mouse received 200 µL RPMI-1640 medium supplemented with Matrigel (Sigma) containing 1 × 105 ES-2 cells (transduced with control or IGF2BP3-targeting shRNA) in the unilateral flank area subcutaneously. The mice were subjected to measurement of tumor volumes every week and monitored for body weight changes and other side effects. Tumor volume was determined by the formula V = 1/2 × L × W2, where L and W separately represent tumor length and width. Mice were euthanized after 30 days to avoid severe discomfort in the control group. Tumor tissues were collected for biochemical or histological assays.
Statistical analysis
Statistical analyses were performed using GraphPad Prism v. 7.0 (GraphPad, CA). The results are reported as means ± standard deviation (SD) from at least three independent experiments. The Student’s t-test was utilized for continuous variables, and statistical significance was determined by a P-value < 0.05.
| Results | ▴Top |
The expression of IGF2BP3 is elevated in OC
To examine the correlation between IGF2BP3 and the advancement of OC, TCGA data of OC tissues were analyzed. TCGA data revealed that IGF2BP3 was notably elevated in the OC tissues compared to the adjacent tissues (Supplementary Material 1, www.wjon.org; Fig. 1a). The single cell sequencing data (GSE154600) revealed that IGF2BP3 expression was significantly higher in the malignant epithelial cells than in the normal epithelial cells from OC tissues (Supplementary Figure 1a, b www.wjon.org). Another GEO data (GSE14407) confirmed that IGF2BP3 was increased in the OC tissues (Supplementary Figure 1c, d, www.wjon.org). Furthermore, the immunoblotting, RT-qPCR, and IHC assay of our collected clinical samples also demonstrated the heightened expression of IGF2BP3 in OC tissues (Fig. 1b-d). Data from Human Protein Atlas database showed enhanced IGF2BP3 protein level in OC tissues (Supplementary Figure 1e, www.wjon.org). Finally, the utilization of Kaplan-Meier plot analysis on OC cohorts demonstrates that OC patients with elevated levels of IGF2BP3 experience a significantly diminished PFS compared to those with lower levels of IGF2BP3 (Fig. 1e). Consequently, these results strongly indicate that the IGF2BP3 is upregulated and closely associated with a detrimental prognosis for OC patients.
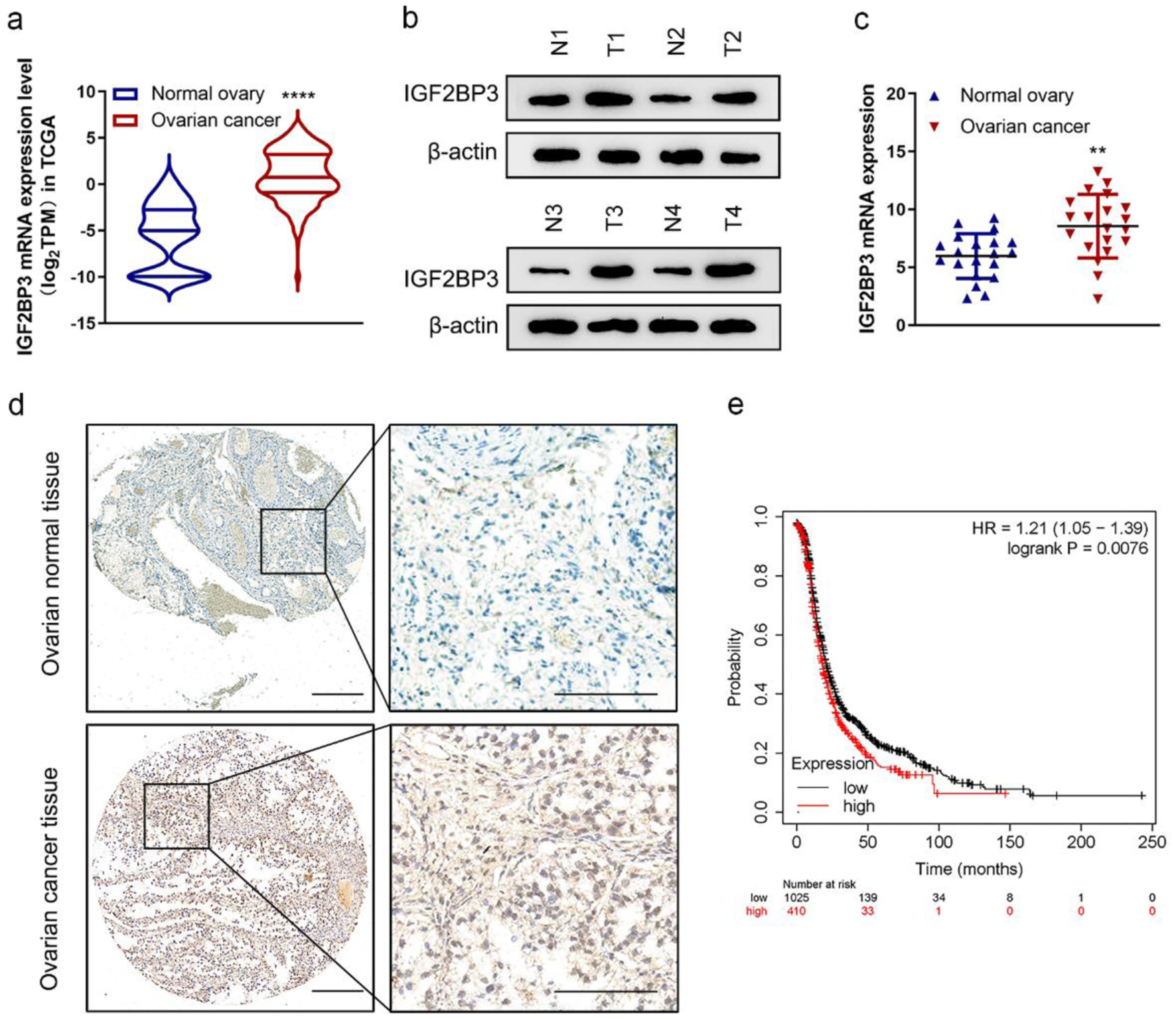 Click for large image | Figure 1. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) expression is increased in ovarian cancer (OC). (a) The mRNA expression of IGF2BP3 in OC (n = 419) tissues versus in the adjacent tissues (n = 88) from The Cancer Genome Atlas (TCGA) data. ****P < 0.0001, OC vs. normal ovary. (b) Representative images of western blot analysis demonstrating the protein level of IGF2BP3 in normal (N) and cancerous (T) ovarian tissues. (c) The expression of IGF2BP3 was analyzed by qRT-PCR and normalized to GAPDH in 20 pairs of normal and cancerous ovarian tissues. (d) Representative immunohistochemistry (IHC) images reflecting the IGF2BP3 expression in OC and normal ovarian tissues. Scale bars: 50 µm (left) and 200 µm (right). (e) Kaplan-Meier curves of progression-free survival (PFS) for OC patients with high (n = 410) and low (n = 1,025) levels of IGF2BP3 (probe ID 203819_s_at). |
Silencing IGF2BP3 inhibits OC cells proliferation
We identified the high IGF2BP3 expression in OC tissues and poorer PFS in OC patients, but IGF2BP3-specific roles remain poorly understood. To explore the functional importance of IGF2BP3 in OC, we analyzed differentially expressed genes in IGF2BP3 knockdown in OC. ES-2 cells were determined (GSE109604). Subsequently, the identified genes were integrated into biological pathways according to their functional context. The result indicated that downregulated genes in IGF2BP3-depleted cells were enriched in cell proliferation process, mRNA stability, and other biological pathways (Fig. 2a). To characterize the functions of IGF2BP2 in OC cells in vitro, two IGF2BP3-targeting shRNAs (sh-IGF2BP3-1 and sh-IGF2BP3-2) were delivered into OC cells (ES-2 and SKOV3) and the short hairpin knockdown efficiency was identified in mRNA and protein level (Supplementary Figure 2a-d, www.wjon.org). The shRNA with higher knockdown efficiency (sh-IGF2PB3-1) was employed for later investigations (sh-IGF2BP3). Cell proliferation by MTS and EdU assays was performed to evaluate the role of IGF2BP3 in OC cell growth. The results revealed that IGF2BP3 inhibition substantially suppressed growth in OC cells (Fig. 2b-g). These results indicate that IGF2BP3 knockdown suppresses OC cell proliferation.
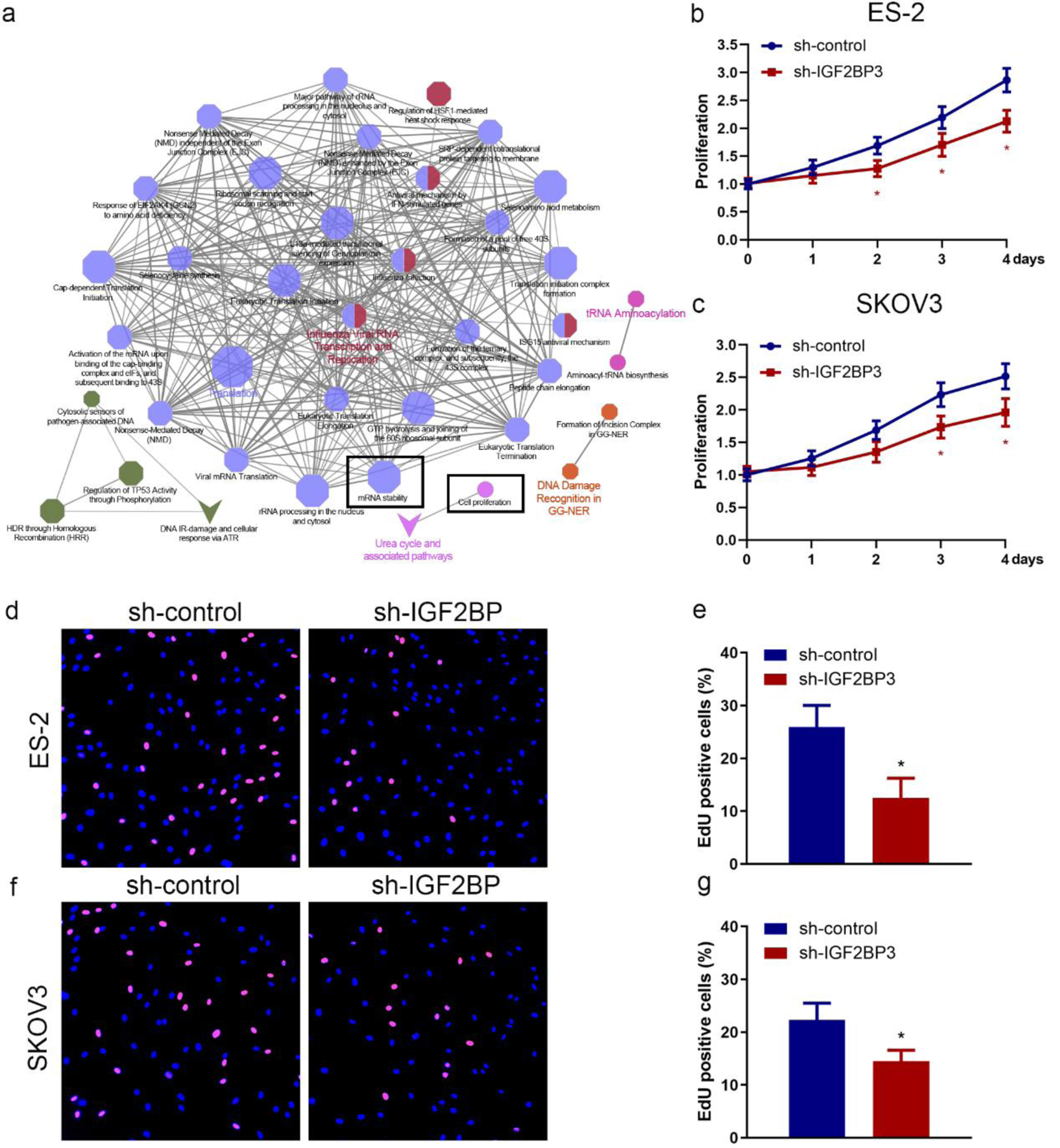 Click for large image | Figure 2. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) knockdown suppresses ovarian cancer (OC) cell proliferation. (a) The pathways impacted by the silencing of IGF2BP3 in OC cells were examined using the clueGO and cluePedia plugins of the cytoscape software. Through this analysis, the cluster of cell proliferation and mRNA stability pathway were found to be enriched and were identified based on statistical analysis. Cell proliferation assay results of MTS assay (b and c) and 5-ethynyl-2′-deoxyuridine (EdU) assay (d-g) evaluating the effect of IGF2BP3 depletion on the growth in ES-2 and SKOV3 cells (*P < 0.05, sh-IGF2BP3 vs. sh-control). (d-g) Representative images (d and f) and quantified results (e and g) of EdU assay determining the effect of IGF2BP3 depletion on the growth in ES-2 (upper panel) and SKOV3 (lower panel) cells. Error bars represent mean ± SD from three experiments. **P < 0.01, sh-IGF2BP3 vs. sh-control. |
IGF2BP3 downregulates PLAGL2 in OC cells and tissues
To elucidate the mechanism by which IGF2BP3 regulates OC cell proliferation, we analyzed the microarray data of IGF2BP3-depleted ES-2 cells (GSE109604). We also explored the genes positively correlated with IGF2BP3 in the TCGA database. Then, we overlapped these two gene sets and identified eight genes positively correlated with IGF2BP3 and downregulated in IGF2BP3-deficient OC cells (Fig. 3a, b). Among these candidate genes, PLAGL2 attracted our attention for several reasons. First, it exhibits a high correlation coefficient with IGF2BP3 with a low P value (Cor = 0.5, P = 4.22 × 10-28, Supplementary Material 2, www.wjon.org); second, PLAGL2 activates the pro-oncogenic signaling, increasing cancer cell proliferation [17]. Nevertheless, how PLAGL2 modulates OC progression remains largely unknown. Meanwhile, bioinformatics analysis (Fig. 3c) and western blotting analysis (Fig. 3d) revealed that PLAGL2 is significantly higher in OC tissues. To investigate whether PLAGL2 expression level changes in tumor tissues, we analyzed PLAGL2 IHC staining was highly expressed in ovarian tissue (Fig. 3e). Moreover, the Kaplan-Meier plot revealed that patients with a high level of PLAGL2 exhibited poor PFS compared with those with low PLAGL2 expression (Fig. 3f). Furthermore, we also noticed that in IGF2BP3-deficient OC cells, the expression of PLAGL2 was downregulated in mRNA and protein levels (Fig. 3g, h). Consistently, the level of IGF2BP3 is identified correlated positively with PLAGL2 in OC tissue (Fig. 3i), which confirmed the positively regulation relationship. These findings further confirmed the correlation between IGF2BP3 and PLAGL2 in OC cells. These data indicate that downregulating IGF2BP3 may suppress the PLAGL2 expression in OC cells and tissues.
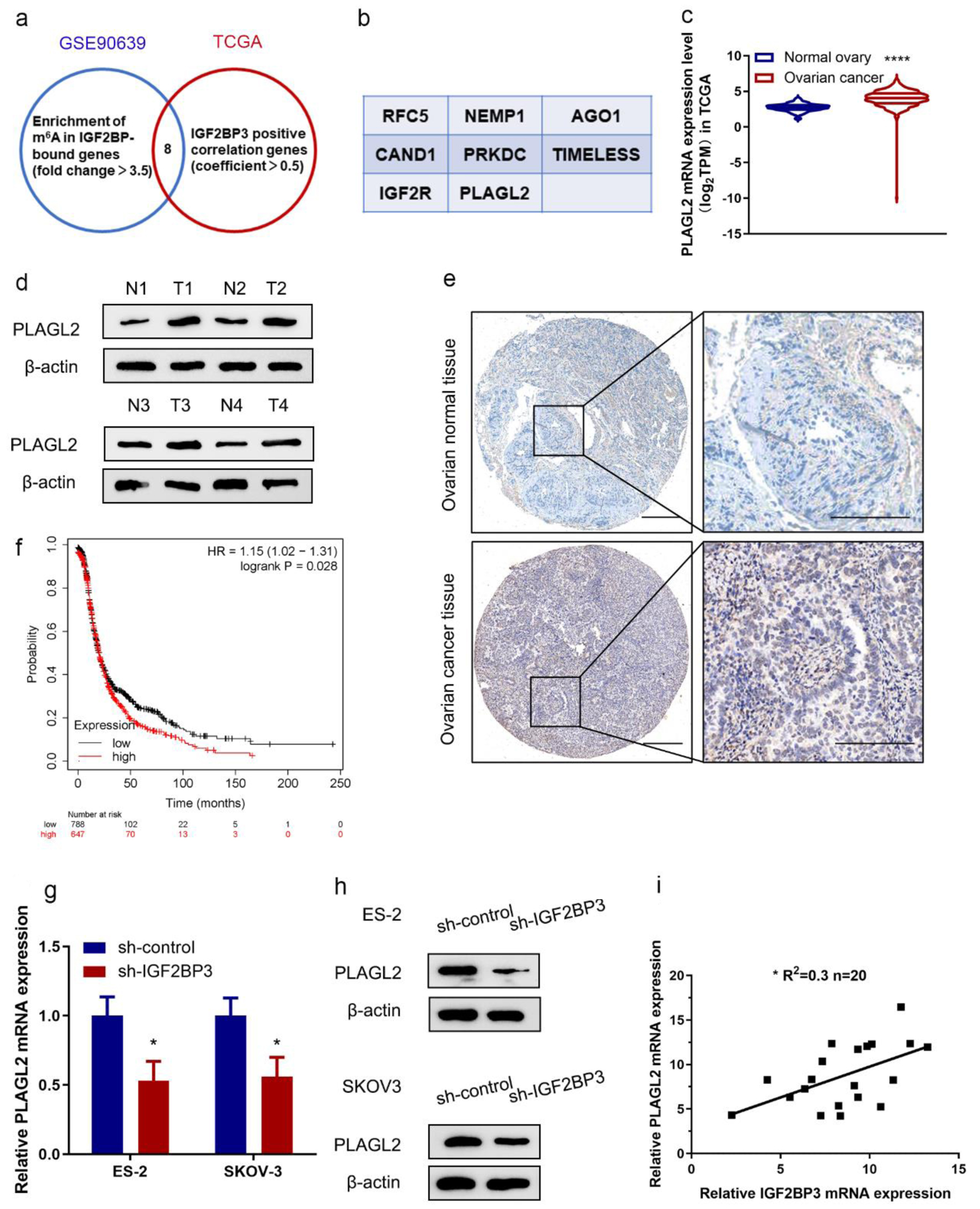 Click for large image | Figure 3. PLAGL2 is a downstream target of insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) in ovarian cancer (OC). (a) Venn diagram showing genes from indicated datasets were positively correlated with IGF2BP3 and/or downregulated in IGF2BP3-deficient OC cells. (b) Genes from the Venn diagram that are both correlated with IGF2BP3 and downregulated in IGF2BP3-dificient OC cells. (c) The mRNA expression of PLAGL2 in OC tissues (n = 419) versus in the adjacent tissues (n = 88) from The Cancer Genome Atlas (TCGA) data. ****P < 0.0001, OC vs. normal ovary. (d) Western blot analysis of PLAGL2 expression in paired normal and cancerous ovarian tissues. (e) Representative immunohistochemistry (IHC) images showing PLAGL2 expression in cancerous and normal ovarian tissues. Scale bars: 50 µm (left) and 200 µm (right). (f) Kaplan-Meier curves of progression-free survival (PFS) for OC patients with high (n = 647) and low (n = 788) levels of PLAGL2 (probe ID 202925_at). (g and h) Results of RT-qPCR (h) and western blot analysis (h) detecting the expression of PLAGL2 in ES-2 and SKOV3 cells transduced with or without IGF2BP3-targeting shRNAs. (i) Paired 20 normal and cancerous ovarian for qRT-PCR assay. Pearson’s correlation analysis of the relative expression levels of IGF2BP3 and PLAGL2. |
Silencing IGF2BP3 attenuates OC cell proliferation in a PLAGL2-dependent manner
As we identified PLAGL2 to be a functional downstream gene of IGF2BP3 in OC cells, we next analyzed the biological role of PLAGL2 in OC cell proliferation by modulating the expression of IGF2BP3. We first upregulated PLAGL2 in parental OC cells (Figure 3, www.wjon.org) and IGF2BP3-depleted cells (Fig. 4a-d) and found that overexpressing PLAGL2 significantly reconstituted the IGF2BP3 knockdown-mediated downregulation of PLAGL2. Then, cell proliferation was investigated, and we found that silencing IGF2BP3 significantly suppressed the OC cell proliferation, while this inhibitory effect was compromised by further overexpressing PLAGL2 (Fig. 4e-h, Supplementary Figure 4, www.wjon.org). This indicated that IGF2BP3 knockdown inhibits OC cell proliferation via downregulating PLAGL2.
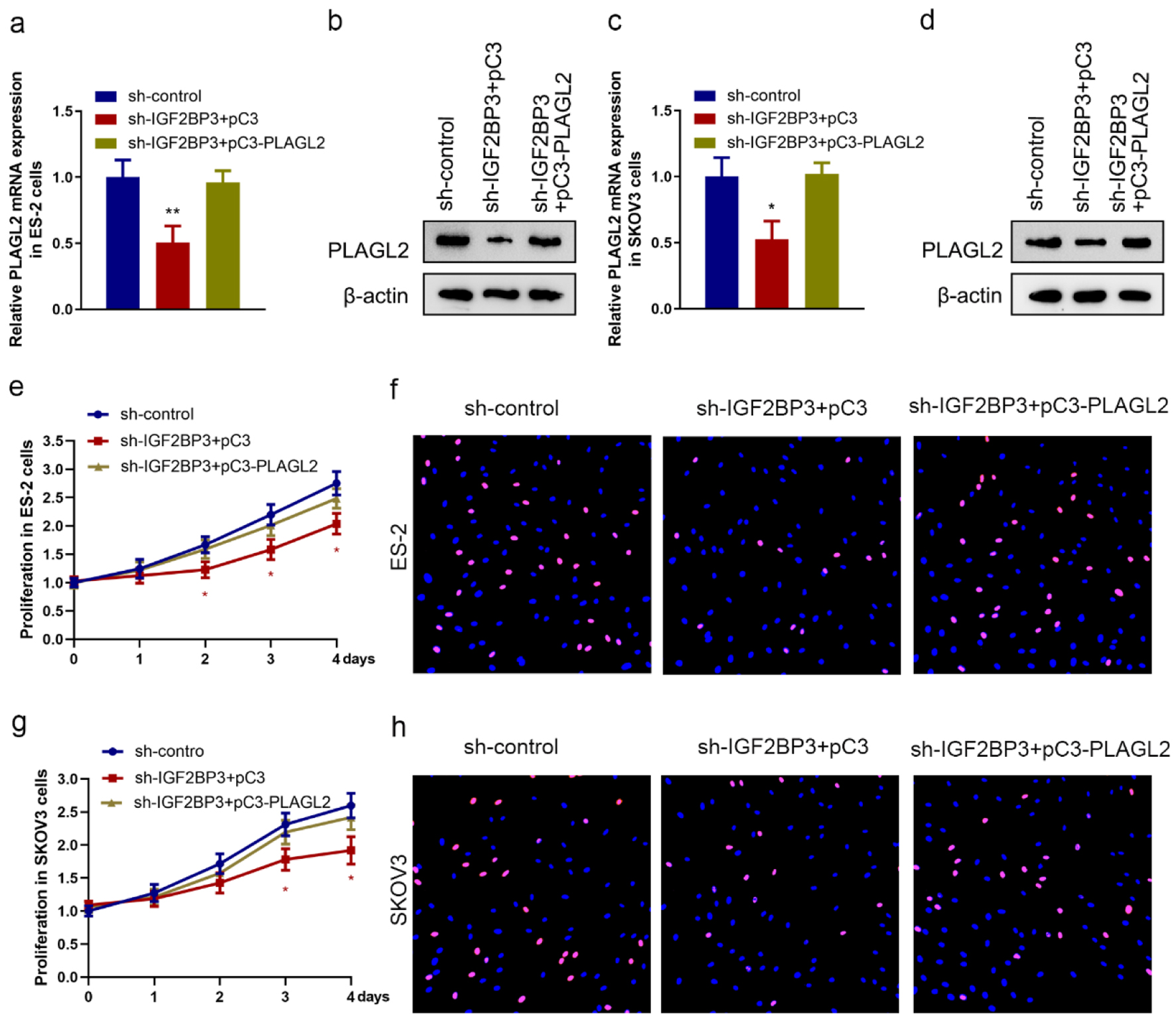 Click for large image | Figure 4. Silencing insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) attenuates ovarian cancer (OC) cell proliferation in a PLAGL2-dependent manner. (a, b, c, and d) The expression of PLAGL2 in IGF2BP3-depleted and PLAGL2 overexpressing cells was assessed using RT-qPCR and western blot analysis in ES-2 cells (a and b) and SKOV3 cells (c and d). (*P < 0.05, **P < 0.01). Cell proliferation results of MTS assays (e and g) and EdU (f and h) assay using ES-2 cells and SKOV3 cells under indicated conditions (IGF2BP3-depleted and PLAGL2 overexpressing cells) (*P < 0.05). |
IGF2BP3 binds PLAGL2 and regulates PLAGL2 expression
IGF2BP3, an mRNA-binding protein, belongs to a distinct family of m6A readers, which effectively targets numerous mRNA transcripts through recognition of the consensus sequence, thereby augmenting both mRNA stability and translation [2]. Using an online bioinformatics tool RBPmap, we found that IGF2BP3 preferentially binds to the m6A modification core motif (Fig. 5a). The binding site of IGF2BP3 on PLAGL2 mRNA was predicted (Fig. 5b). Next, we sought to investigate whether IGF2BP3 interacted with PLAGL2 mRNA. Consistent with the bioinformatics, our RIP-qPCR assays demonstrated IGF2BP3 could precipitated with the mRNA of PLAGL2 in ES-2 cells (Fig. 5c). The presence of IGF2BP3 within the PLAGL2 sense RNA probe pull-down cell samples in ES-2 cells was confirmed through western blot analysis (Fig. 5d). Then, to test the impact of IGF2BP3 on PLAGL2 mRNA stability, we determined the PLAGL2 half-life mRNA in control or IGF2BP3-depleted OC cells. We found accelerated PLAGL2 mRNA decay in IGF2BP3-deficient ES-2 cells (Fig. 5e). IGF2BP3 knockdown resulted in the inhibition of exogenous mRNA decay, as evidenced by a decrease in the half-life of PLAGL2 mRNA from 62 to 40 min (Fig. 5f). These data suggested that IGF2BP3 bound to PLAGL2 mRNA and increased its stability.
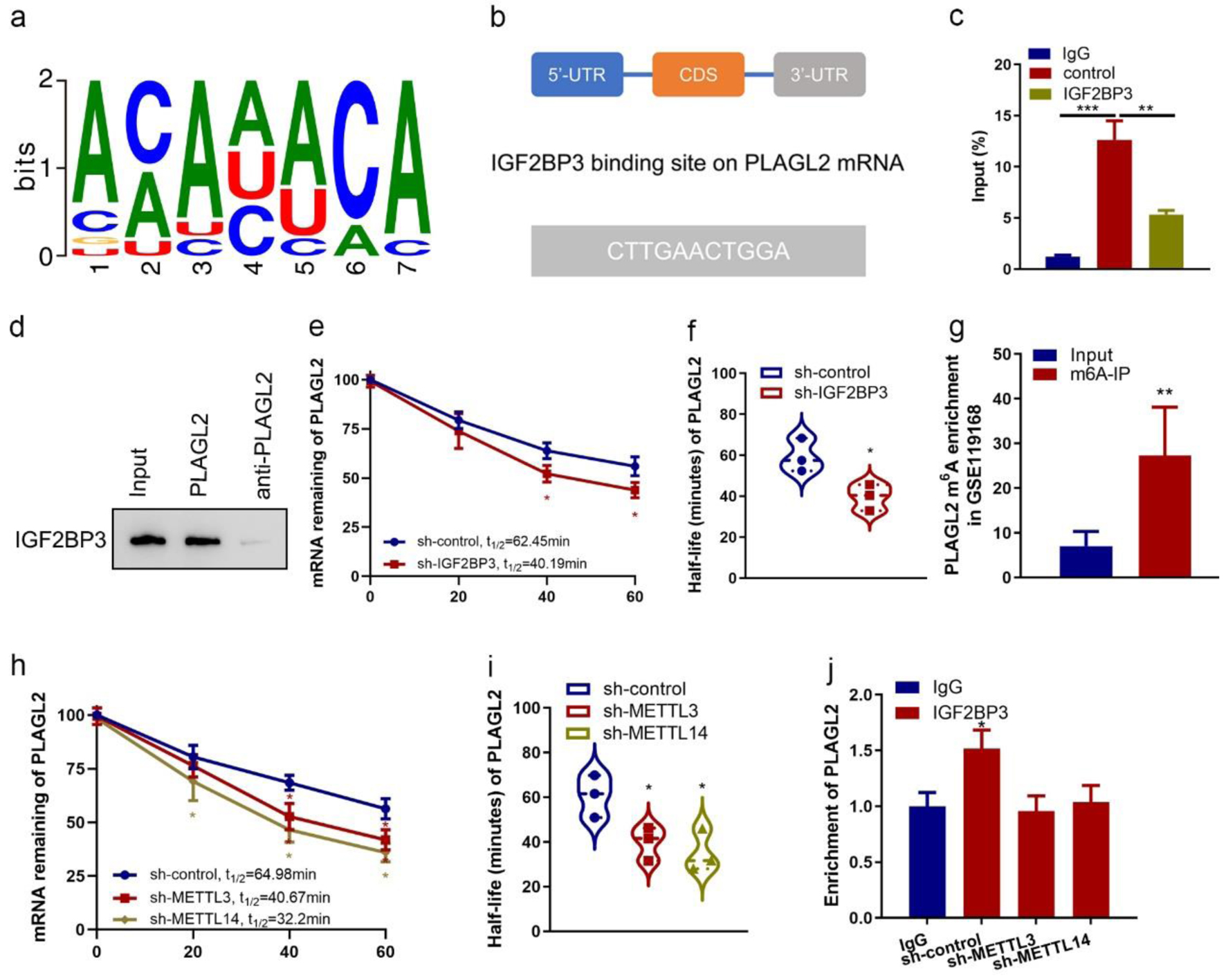 Click for large image | Figure 5. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) binds with PLAGL2 mRNA and regulates its stability. (a) Consensus sequences of IGF2BP3-binding sites by motif prediction. (b) The binding site of IGF2BP3 on PLAGL2 mRNA was predicted. The 3′-UTR sequence with potential binding sites (blue) is shown. (c) Enrichment of PLAGL2 in ES-2 cells performed RIP of IGF2BP3 and control using anti-flag antibody from nuclear extractions and identified the associated PLAGL2 mRNAs by RT-qPCR. (***P < 0.001, **P < 0.01). (d) The cell lysate of ES-2 cells was utilized in an RNA pulldown assay employing biotin-labeled PLAGL2 and PLAGL2 antisense, followed by western blotting analysis. (e) Result of the mRNA half-life measurement showing the decaying of PLAGL2 mRNA in parental and IGF2BP3-depleted cells (*P < 0.05, sh-IGF2BP3 vs. sh-control). (f) Half-life values in the same treatment groups. (g) Results of bioinformatics analysis of online dataset (GSE90639) revealing the enrichment of PLAGL2 m6A-modified RNA by IGF2BP3 (***P < 0.001, m6A probe vs. unmethylated probe). (h) Result of the mRNA half-life measurement showing the decaying of PLAGL2 mRNA in parental and METTL3- or METTL14-depleted cells (*P < 0.05, vs. sh-control). (i) Half-life values in the same treatment groups. (j) Enrichment of PLAGL2 in parental and METTL3- or METTL14-depleted ES-2 cells performed RIP of IGF2BP3 and control using anti-flag antibody and identified the associated PLAGL2 mRNAs by RT-qPCR. Input percentage is shown (*P < 0.05). |
We next want to know how IGF2BP3 regulated PLAGL2 mRNA stability. Recent studies revealed that m6A modifications could enhance or disrupt RNA stability, which depends on the m6A readers’ specificity, through dynamic interplays with RNA-binding proteins [18]. So, we evaluated whether the IGF2BP3 binding to the PLAGL2 mRNA is associated with m6A modification. Data from the GEO dataset (GSE119168) of clinical patients’ samples showed that m6A-modified PLAGL2 mRNAs in the OC tissues were much higher than in the normal tissues (Fig. 5g). Additionally, the silencing of PLAGL2 m6A writers, METTL3 and METTL14, also reduces the level and half-life of PLAGL2 mRNA (Fig. 5h, i, Figure 5, www.wjon.org). RIP assay showed that METTL3 or METTL14 knockdown also reduced PLAGL2 enrichment in the anti-IGF2BP3 immunoprecipitates in the ES-2 cells (Fig. 5j). These findings revealed that IGF2BP3 regulated PLAGL2 mRNA stability and their direct interaction was dependent on m6A modification.
m6A modification mediates stabilization of PLAGL2 mRNA by IGF2BP3
To further explore the m6A mRNA mechanism that IGF2PB3 regulates PLAGL2 mRNA stability in OC, we predicted the m6A modification site in PLAGL2 transcripts using an online tool m6AVar. The results revealed a potential m6A site located in the 3'-UTR 979-989 bp region with very high confidence (Fig. 6a). Using PLAGL2-specific m6A RT-qPCR assay, this region was confirmed as an m6A modification site (Fig. 6b). Meanwhile, RIP assay also showed that PLAGL2 3'-UTR could be immunoprecipitated by METTL3 and METTL14, respectively (Fig. 6c), indicating the involvement of these writers in the PLAGL2 m6A modification. To further illustrate the molecule mechanism, we generated reporter plasmids expressing PLAGL2 3'-UTR containing wildtype or mutated m6A site (Fig. 6d) and performed a dual luciferase reporter assay. We observed that IGF2BP3 significantly increased the luciferase activity in cells expressing wildtype 3'-UTR of PLAGL2 but not in those expressing mutated PLAGL2 3'-UTR (Fig. 6e). Conversely, both silence of IGF2BP3 and m6A writer METTL14 inhibited the luciferase activity with wildtype m6A modification site of PLAGL2 3’-UTR but failed to suppress the luciferase activity with mutant m6A modification site of PLAGL2 3’-UTR (Fig. 6f, g). Moreover, silence of the m6A writer METTL14 completely abrogated the m6A reader IGF2BP3-induced upregulation of luciferase activity with wildtype m6A modification site of PLAGL2 (Fig. 6h). Altogether, these results suggested that m6A modification mediated upregulation of PLAGL2 through corporation of IGF2BP3 and METTL14.
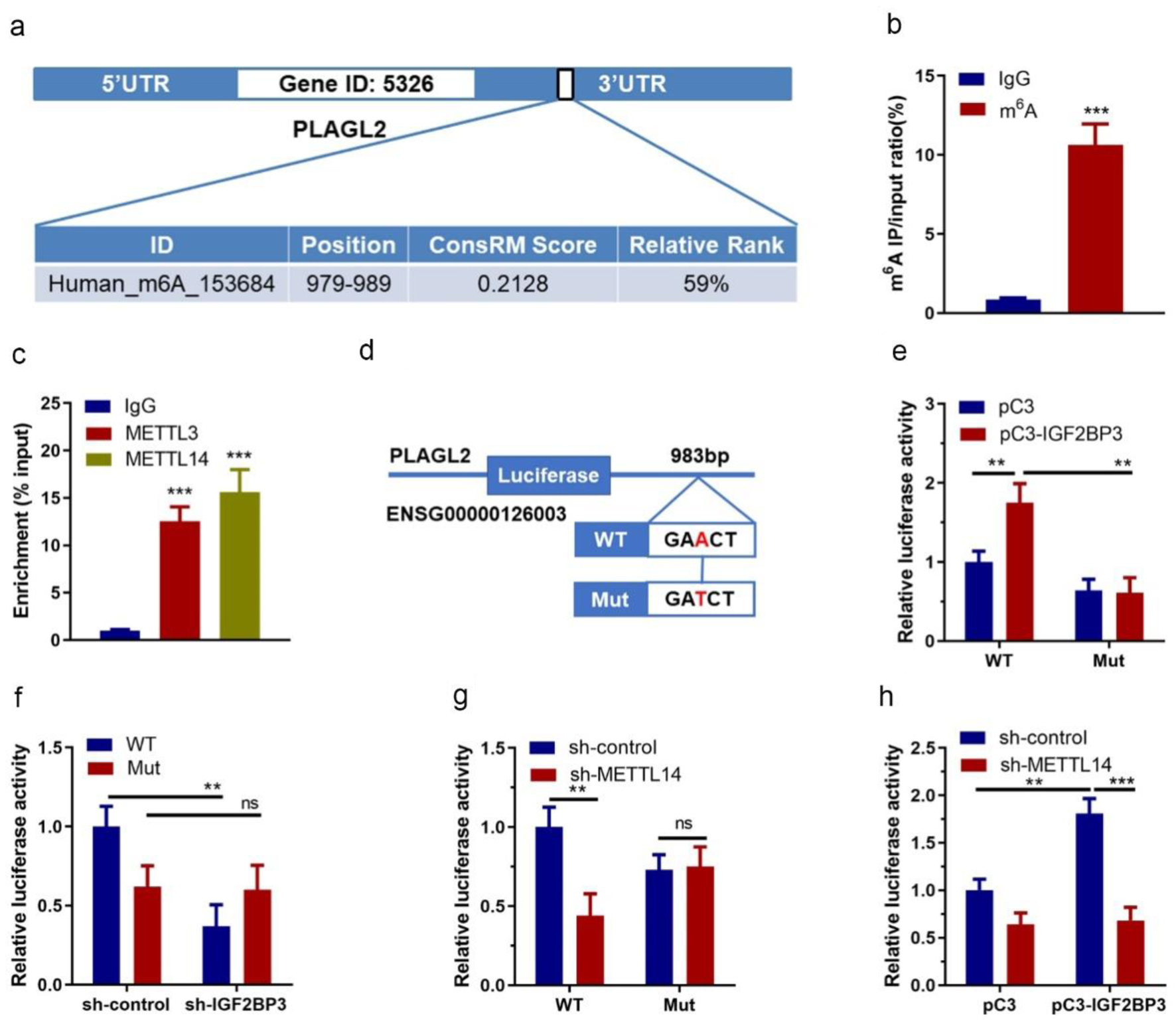 Click for large image | Figure 6. Identification of m6A methylation between PLAGL2 mRNA and insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3). (a) The prediction of the PLAGL2 m6A RNA methylation site using online tool (m6Avar). (b) Results of PLAGL2-specific m6A RT-qPCR assay (*** P<0.001, m6A vs. IgG). (c) Results of RIP assay showing the interactions between PLAGL2 and METTL3/METTL14 (***P < 0.001, vs. IgG). (d) A schematic diagram showing the wildtype and mutated methylation site on PLAGL2 3'-UTR. (e) Results of dual luciferase assay in ES-2 cells co-transfected with vechicle vector or IGF2BP3-overexpressing vector and reporter plasmid containing wildtype or mutant PLAGL2 3'-UTR (*P < 0.05, **P < 0.01, vs. pC3). (f) Results of dual luciferase assay in ES-2 cells with or without IGF2BP3 knockdown transfected with reporter plasmid containing wildtype or mutant PLAGL2 3'-UTR (*P < 0.05, vs. WT sh-control). (g) Results of dual luciferase assay in ES-2 cells co-transfected with vechicle vector or METTL14-targeting shRNA and reporter plasmid containing wildtype or mutant PLAGL2 3'-UTR (*P < 0.05, vs. WT sh-control). (h) Relative luciferase activity of PLAGL2 wildtype 3'-UTR or mutation in METTL14 stable knockdown or control ES-2 cells with ectopic expression of IGF2BP3. |
IGF2BP3 knockdown impairs tumor growth in vivo
To examine the biological effect of IGF2BP3 on OC progression in vivo, we generated a subcutaneous xenograft model using ES-2 cells with stable knockdown of IGF2BP3 or control in nude mice. The in vivo data revealed that silencing IGF2BP3 significantly inhibited the tumor growth in nude mice bearing human OC xenografts compared with the sh-control, as tumor tissues excised from nude mice shown (Fig. 7a). Meanwhile, the tumor volume and weight were decreased in IGF2BP3 knockdown ES-2 cells xenografts (Fig. 7b, c). In addition, reduced expression of IGF2BP3 and PLAGL2 at transcriptional and translational levels were confirmed in xenografts tissues (Fig. 7d, e). These data above suggested a vital role of IGF2BP3 in OC growth in vivo.
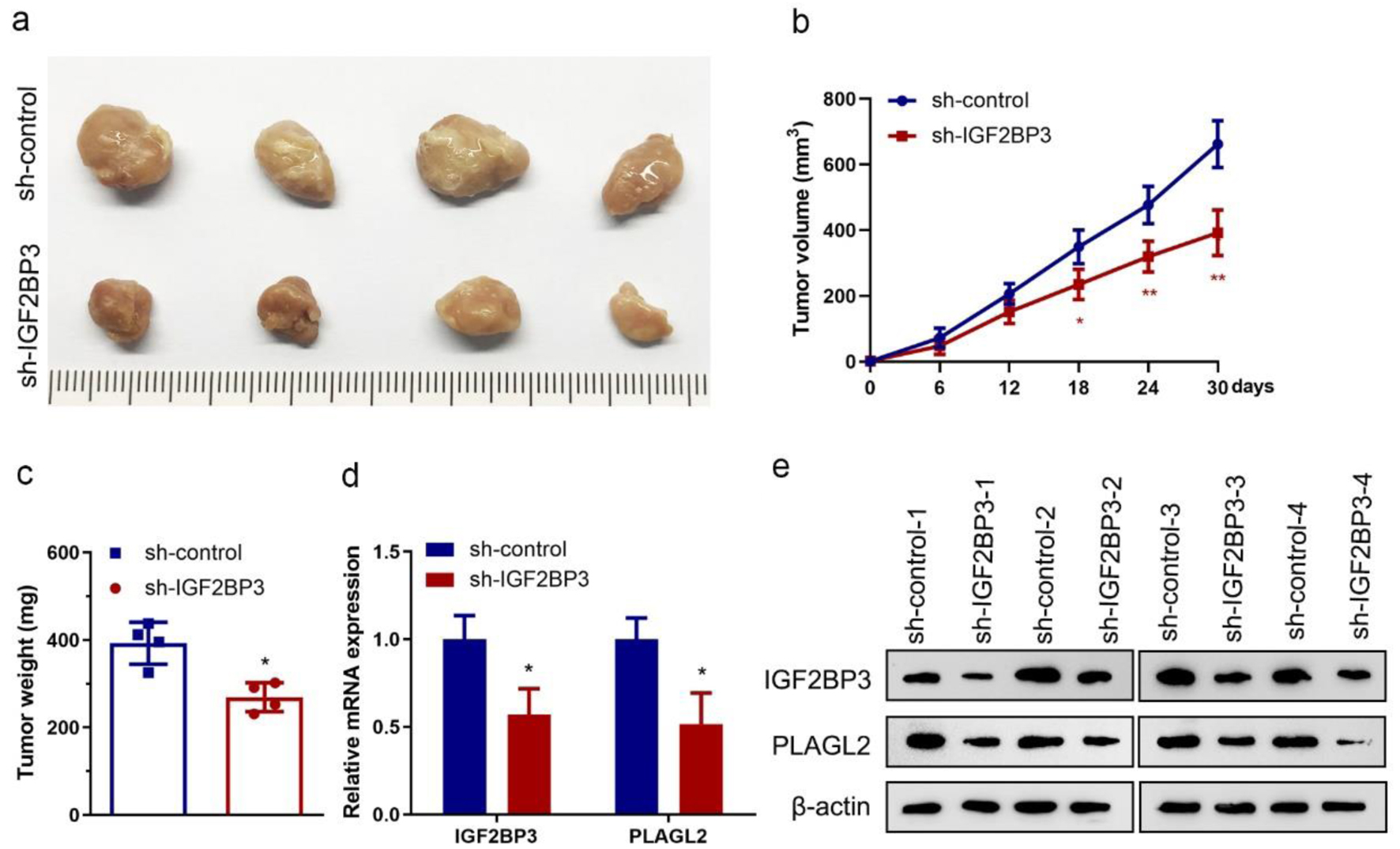 Click for large image | Figure 7. Insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) knockdown impairs tumor growth in vivo. (a) Images showing tumors harvested from each group. (b) Plotted data depicting tumor growth curves in different groups (n = 4) (*P < 0.05, **P < 0.01, vs. sh-control). (c) Analyzed data showing the individual tumor weight from each group (n = 4) (*P < 0.05, vs. sh-control). (d and e) Results of RT-qPCR (d) and representative images of western blot analysis (e) showing the expression of IGF2BP3 and PLAGL2 in harvested xenograft tumors (*P < 0.05, vs. sh-control). |
| Discussion | ▴Top |
The unregulated IGF2BP3 in tumors and different cancers indicates IGF2BP3 serves oncogenic roles. However, as an m6A reader, the sets of significantly IGF2BP3 depletion caused by dysregulated biological pathways in OC are unknown. In mechanism research, IGF2BP3 could bind to 3'-UTR of PLAGL2 to maintain the mRNA stability. In in vivo experiments, IGF2BP3 knockdown suppressed OC cell proliferation via inhibiting PLAGL2 expression. In summary, our results showed that PLAGL2 mediates the main function of IGF2BP3 knockdown on OC proliferation inhibition through mRNA stability regulation (Fig. 8).
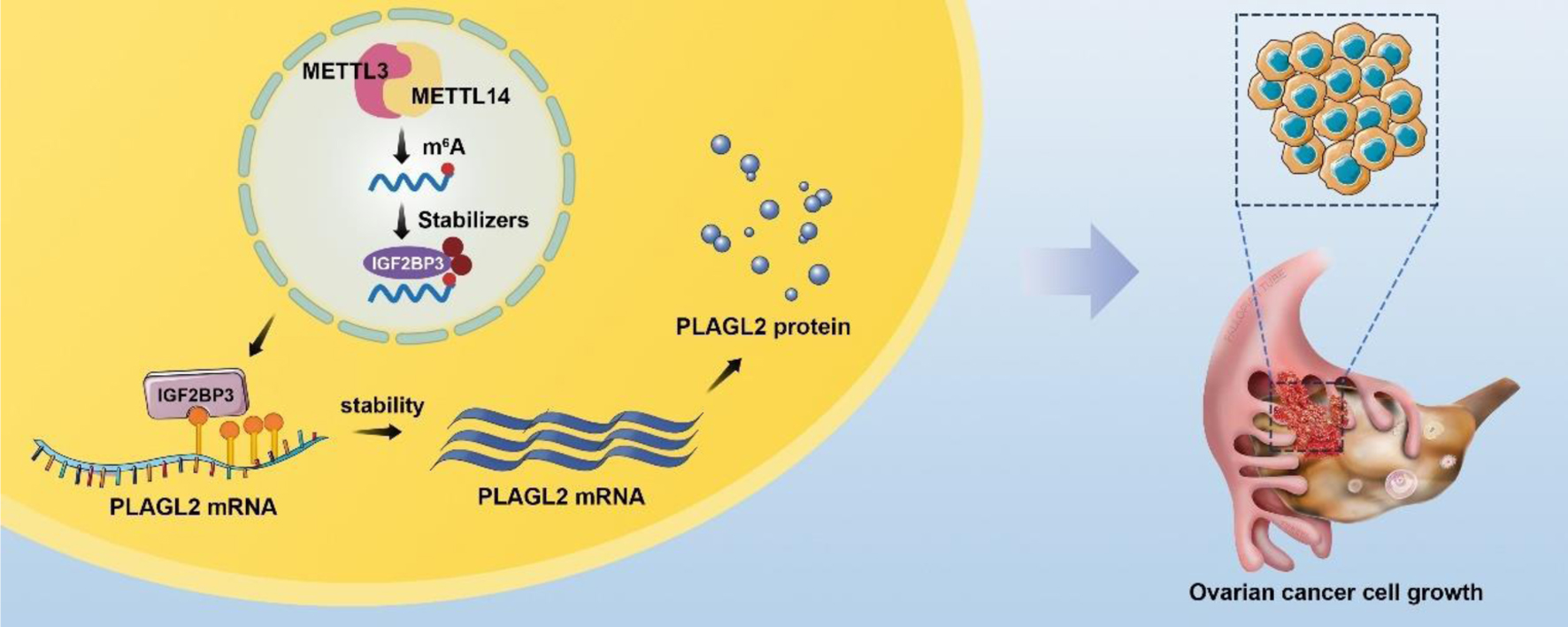 Click for large image | Figure 8. A schematic diagram for the mechanisms of insulin-like growth factor 2 mRNA-binding protein family (IGF2BP3) knockdown on ovarian cancer proliferation inhibition is mediated by PLAGL2 through mRNA stability regulation. In mechanism research, knockdown IGF2BP1 inhibits cell proliferation and is downstream gene PLAGL2 mRNA stability. Like IGF2BP1, unregulated PLAGL2 indicates a poor prognosis in ovarian cancer. Together this indicates that IGF2BP1 knockdown represses ovarian cancer cell proliferation largely by regulating target gene PLAGL2 mRNAs stability. |
In this study, by analyzing the RNA-seq results obtained from the TCGA database, we identified genes positively correlated with IGF2BP3 and found PLAGL2 was the most promising target gene. Meanwhile, we explored the genes in GEO dataset (GSE109604) and found PLAGL2 mRNA was pulled down by IGF2BP3. Subsequently, we identified an IGF2BP3 m6A-dependent binding site on PLAGL2 3'-UTR. Based on these observations, we speculated that IGF2BP3 may recognize the PLAGL2 mRNA and regulate its stability. A recent study reported that m6A writer METTL14 promotes the MYC mRNA stability, in which IGF2BP3-mediated-m6A-dependent mechanism may also involve [2]. In our study, we also found that PLAGL2 mRNA stability was maintained by the IGF2BP3-mediated-m6A-dependent mechanism. More importantly, to the best of our knowledge, our study was the first to report that IGF2BP3-involved m6A methylation mediated stabilization of PLAGL2 and regulated OC growth.
The involvement of RBP in tumor progression and the underlying mechanisms have been documented in recent years [19]. Among these reported RBPs, IGF2BP3 has been recognized as an oncogene in various types of cancers including endometrial carcinoma and nasopharyngeal carcinoma [19, 20]. IGF2BP3 has been implicated in tumor metastasis and unfavorable survival outcomes among patients with nasopharyngeal carcinoma [20]. Furthermore, elevated levels of IGF2BP3 have been observed to correlate with poorer overall survival and serve as a potential prognostic marker for lung adenocarcinoma [21]. Additionally, a prior investigation has provided evidence that IGF2BP3 facilitates the advancement of endometrial carcinoma by augmenting the mRNA stability of E2F3 [19]. Moreover, in the context of bladder cancer, IGF2BP3 has been observed to stimulate cellular proliferation and progression through the cell cycle, thereby facilitating tumorigenesis [22]. In recent research, it was discovered that the m6A “reader” IGF2BP3 exhibits the ability to recognize m6A-modified mRNAs and bolster their stability [2]. In line with these previous studies, we have shown that IGF2BP3 interacts with m6A-modified PLGAL2 mRNA at the consensus sequence to stabilize it to promote the tumor growth. Meanwhile, silencing IGF2BP3 facilitated the decaying of PLGAL2 mRNA, which ultimately suppressed the tumor cell proliferation. Consistently, a recent study shows that IGF2BP3 recognizes the consensus GG(m6A) and binds to their target mRNAs, including MYC and TMBIM6 [2, 23].
The carcinogenic effects of PLAGL2 have been observed in numerous malignancies and are commonly upregulated in various types of cancer, including OC and colorectal cancer [24-26]. PLAGL2 is responsible for activating pro-oncogenic signaling pathways, consequently enhancing cancer cell proliferation, migration, and metastasis [25, 26]. Notably, PLAGL2 has been demonstrated to facilitate cell proliferation in gastric cancer [17], and targeting PLAGL2 has been proven to effectively suppress tumor proliferation and metastasis in colorectal cancer [27]. Under the scenario of OC, PLAGL2 shows an oncogenic role in enhancing ovarian tumorigenesis and promoting malignant phenotypes of OC cells [25, 28]. These findings are consistent with our observations that elevated PLAGL2 is detected in cancerous ovarian tissues and is correlated with a negative prognosis for patients with ovarian cancer. Furthermore, our in vitro and in vivo data from the current study indicate that decreased PLGAL2 is linked to inhibited cancer cell proliferation and diminished tumor growth, which is substantiated by the abovementioned studies.
In this study, we have presented evidence supporting the significant involvement of disrupting IGF2BP3-mediated m6A modification in OC cell proliferation. Our findings suggest that the silencing of IGF2BP3 leads to a reduction in m6A methylation modification of PLAGL2, consequently compromising the mRNA stability of PLAGL2 in OC. Furthermore, our future investigations indicate that targeting IGF2BP3-dependent networks may hold promise as a therapeutic strategy for OC.
| Supplementary Material | ▴Top |
Suppl 1. Correlation analysis of the relative genes and IGF2BP3.
Suppl 2. mRNA raw data of PLAGL2 and IGF2BP3 in TCGA database.
Suppl 1. IGF2BP3 expression of ovarian cancer was confirmed by data from GEO and Human Protein Atlas database. (a) Analysis of single-cell transcriptome data (GSE154600) in human OC tissue. (b) IGF2BP3 expression between normal epithelial cell and malignant epithelial cells was tested by single cell sequencing and compared. (c) IGF2BP3 expression of ovarian cancer in GEO data of GSE14407. (d) Raw data of IGF2BP3 RNA levels in GEO database (Accession GSE14407, dataset GDS3592, gene ID: 203820_s_at). (e) Comparison of relative IGF2BP3 expression between ovarian cancer and normal tissues in The Human Protein Atlas database.
Suppl 2. IGF2BP3 lentiviral shRNA knockdown efficacy identification. Data from RT-qPCR (a and c) and western blot (b and d) analyses showing the downregulation of IGF2BP3 in ES-2 (left) and SKOV3 (right) cells infected with lentiviral expressing control or IGF2BP3-targeting shRNAs. β-actin was used as an internal loading control (*P < 0.05, **P < 0.01, sh-IGF2BP3s vs. pC3).
Suppl 3. PLAGL2 lentiviral overexpression efficacy identification. Data from RT-qPCR (a and c) and western blot (b and d) analyses showing the upregulation of PLAGL2 in ES-2 (left) and SKOV3 (right) cells infected with lentiviral expressing control or PLAGL2 -overexpression lentiviral. β-actin was used as an internal loading control (*P < 0.05, **P < 0.01, pC3-PLAGL2 vs. pC3).
Suppl 4. Silencing IGF2BP3 attenuates OC cell proliferation. Quantitative analysis showing clone formation in IGF2BP3-depleted and PLAGL2 overexpressing cells in ES-2 cells (a) and SKOV3 cells (b) (*P < 0.05).
Suppl 5. METTL3 and METTL14 lentiviral knockdown efficacy identification. Data from RT-qPCR (a and c) and western blot (b and d) analyses showing the downregulation of METTL3 and METTL14 in ES-2 cells infected with lentiviral expressing shRNAs. The β-actin was employed as the internal control (*P < 0.05, sh-control vs. sh-METTL3, sh-control vs. sh-METTL14).
Acknowledgments
None to declare.
Financial Disclosure
This study was supported by grants from by the National Natural Science Foundation of China (82373343 to WB and 81702554 to YL).
Conflict of Interest
None to declare.
Informed Consent
Informed consent was obtained from all participants.
Author Contributions
Experiments: Tiantian Dai and Yize Li; acquisition of patient samples: Huiting Hu and Hongyan Peng; analysis and interpretation of data: Yongmei Zhao; financial support: Wendong Bai; Study design: Wendong Bai; drafting of the manuscript: Tiantian Dai and Yize Li; revised of the manuscript: Wendong Bai and Jingwen Wang.
Data Availability
All data that support the findings of this study are available to the corresponding authors by reasonable request.
Abbreviations
ATCC: American Type Culture Collection; EdU: 5-ethynyl-2′-deoxyuridine; IGF2BP3: insulin-like growth factor 2 mRNA-binding protein family; IHC: immunohistochemistry; METTL3: methyltransferase like protein 14; METTL14: methyltransferase like protein 14; OC: ovarian cancer; PFS: progression-free survival; PLAGL2: PLAG1 like zinc finger 2; RIP: RNA immunoprecipitations; RT-qPCR: quantitative real-time PCR analysis; TCGA: The Cancer Genome Atlas
| References | ▴Top |
- Chen LJ, Liu HY, Xiao ZY, Qiu T, Zhang D, Zhang LJ, Han FY, et al. IGF2BP3 promotes the progression of colorectal cancer and mediates cetuximab resistance by stabilizing EGFR mRNA in an m(6)A-dependent manner. Cell Death Dis. 2023;14(9):581.
doi pubmed pmc - Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285-295.
doi pubmed pmc - Li Y, Peng H, Jiang P, Zhang J, Zhao Y, Feng X, Pang C, et al. Downregulation of methyltransferase-like 14 promotes ovarian cancer cell proliferation through stabilizing TROAP mRNA. Front Oncol. 2022;12:824258.
doi pubmed pmc - Shen J, Tao YJ, Guan H, Zhen HN, Dong TT, Liu ZK, Zhang FQ. Abdominopelvic lymphatic drainage area irradiation for consolidative radiotherapy of advanced ovarian carcinoma: analysis of clinical application efficacy and dosimetric verification. World J Oncol. 2022;13(3):145-154.
doi pubmed pmc - Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, Guo W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21(1):60.
doi pubmed pmc - Samuels TJ, Jarvelin AI, Ish-Horowicz D, Davis I. Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. Elife. 2020;9:e51529.
doi pubmed pmc - http://cancergenome.nih.gov/
- http://www.proteinatlas.org/
- http://rbpmap.technion.ac.il
- http://m6avar.renlab.org/index.html
- http://kmplot.com/analysis/
- Taha NA, Shafiq AM, Mohammed AH, Zaky AH, Omran OM, Ameen MG. FOS-like antigen 1 expression was associated with survival of hepatocellular carcinoma patients. World J Oncol. 2023;14(4):285-299.
doi pubmed pmc - Warli SM, Mantiri BJ, Sihombing B, Siregar GP, Prapiska FF. Nephrolithiasis-associated renal cell carcinoma in patients who underwent nephrectomy: a single-center experience. World J Oncol. 2023;14(1):94-100.
doi pubmed pmc - Chen YD, Gao KX, Wang Z, Deng Q, Chen YT, Liang H. Glycine decarboxylase suppresses the renal cell carcinoma growth and regulates its gene expressions and functions. World J Oncol. 2022;13(6):387-402.
doi pubmed pmc - Min WL, Wang BF, Liang BB, Zhang L, Pan JY, Huang Y, Zhao Y, et al. A ROS/Akt/NF-kappaB signaling cascade mediates epidermal growth factor-induced epithelial-mesenchymal transition and invasion in human breast cancer cells. World J Oncol. 2022;13(5):289-298.
doi pubmed pmc - Wang YM, Peng ZY, Zhang LY, Wang YX, Fan RH, Zhang H, Jiang WH. N6-methyladenosine RNA modification landscape in the occurrence and recurrence of nasopharyngeal carcinoma. World J Oncol. 2022;13(4):205-215.
doi pubmed pmc - Wu L, Zhao N, Zhou Z, Chen J, Han S, Zhang X, Bao H, et al. PLAGL2 promotes the proliferation and migration of gastric cancer cells via USP37-mediated deubiquitination of Snail1. Theranostics. 2021;11(2):700-714.
doi pubmed pmc - Chen H, Wang Y, Su H, Zhang X, Chen H, Yu J. RNA N(6)-methyladenine modification, cellular reprogramming, and cancer stemness. Front Cell Dev Biol. 2022;10:935224.
doi pubmed pmc - Wang C, Kong F, Ma J, Miao J, Su P, Yang H, Li Q, et al. IGF2BP3 enhances the mRNA stability of E2F3 by interacting with LINC00958 to promote endometrial carcinoma progression. Cell Death Discov. 2022;8(1):279.
doi pubmed pmc - Xu Y, Guo Z, Peng H, Guo L, Wang P. IGF2BP3 promotes cell metastasis and is associated with poor patient survival in nasopharyngeal carcinoma. J Cell Mol Med. 2022;26(2):410-421.
doi pubmed pmc - Guo W, Huai Q, Wan H, Guo L, Song P, Gao S, He J. Prognostic impact of IGF2BP3 expression in patients with surgically resected lung adenocarcinoma. DNA Cell Biol. 2021;40(2):316-331.
doi pubmed - Huang W, Li Y, Zhang C, Zha H, Zhou X, Fu B, Guo J, et al. IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer. J Cell Mol Med. 2020;24(23):13949-13960.
doi pubmed pmc - Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, Li Q, et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021;40(1):80.
doi pubmed pmc - Landrette SF, Kuo YH, Hensen K, Barjesteh van Waalwijk van Doorn-Khosrovani S, Perrat PN, Van de Ven WJ, Delwel R, et al. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood. 2005;105(7):2900-2907.
doi pubmed - Li C, Dong B, Xu X, Li Y, Wang Y, Li X. LncRNA ARAP1-AS1 aggravates the malignant phenotypes of ovarian cancer cells through sponging miR-4735-3p to enhance PLAGL2 expression. Cytotechnology. 2021;73(3):363-372.
doi pubmed pmc - Li N, Li D, Du Y, Su C, Yang C, Lin C, Li X, et al. Overexpressed PLAGL2 transcriptionally activates Wnt6 and promotes cancer development in colorectal cancer. Oncol Rep. 2019;41(2):875-884.
doi pubmed pmc - Zhou Z, Wu L, Liu Z, Zhang X, Han S, Zhao N, Bao H, et al. MicroRNA-214-3p targets the PLAGL2-MYH9 axis to suppress tumor proliferation and metastasis in human colorectal cancer. Aging (Albany NY). 2020;12(10):9633-9657.
doi pubmed pmc - Majem B, Parrilla A, Jimenez C, Suarez-Cabrera L, Barber M, Marin A, Castellvi J, et al. MicroRNA-654-5p suppresses ovarian cancer development impacting on MYC, WNT and AKT pathways. Oncogene. 2019;38(32):6035-6050.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


