| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 2, April 2024, pages 257-267
High Ki67 Gene Expression Is Associated With Aggressive Phenotype in Hepatocellular Carcinoma
Vicente Ramos-Santillana, g, Masanori Oshia, b, g, Erek Nelsona, Itaru Endob, Kazuaki Takabea, b, c, d, e, f, h
aDepartment of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
bDepartment of Surgery, Yokohama City University, Yokohama, Japan
cDivision of Surgical Oncology, Department of Surgery, Virginia Commonwealth University School of Medicine and Massey Cancer Center, Richmond, VA, USA
dDepartment of Surgery, University at Buffalo Jacob School of Medicine and Biomedical Sciences, the State University of New York, Buffalo, NY, USA
eDepartment of Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
fDepartment of Breast Surgery and Oncology, Tokyo Medical University, Tokyo, Japan
gThese authors contributed equally to this work.
hCorresponding Author: Kazuaki Takabe, Department of Surgical Oncology, Roswell Park Cancer Institute, Buffalo, NY 14263, USA
Manuscript submitted October 28, 2023, accepted December 20, 2023, published online March 21, 2024
Short title: High MKi67 Expression and Aggressive Biology in HCC
doi: https://doi.org/10.14740/wjon1751
| Abstract | ▴Top |
Background: Hepatocellular carcinoma (HCC) with high Ki67 protein expression, the most commonly used cell proliferation marker, is associated with an aggressive biologic phenotype; however, conventional immunostaining is hampered by variability in institutional protocol, specific antibody probe, and by assessor subjectivity. To this end, we hypothesized that Ki67 gene (MKi67) expression would identify highly proliferative HCC, and clarify its association with oncologic outcome, tumor progression, and immune cell population in the tumor microenvironment (TME). Furthermore, we sought to identify the cell-cycle gene expression profile that confers this aggressive phenotype.
Methods: A total of 473 HCC patients with clinicopathological data associated with transcriptome were selected for this study: 358 patients from The Cancer Genome Atlas (TCGA) as the testing cohort, and 115 from GSE76427 as the validation cohort. Each cohort was divided into a highly proliferative group (MKi67-high) and the low MKi67 group (MKi67-low) by the median of Ki67 gene (MKi67) expression levels.
Results: MKi67-high HCC patients had worse disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) independent of histological grade in the TCGA cohort. MKi67 expression correlated with histological grade and tumor size. MKi67 expression increased throughout the HCC carcinomatous sequence from normal liver, cirrhotic liver, early HCC, and advanced HCC. MKi67-high HCC was associated with higher intratumor heterogeneity, homologous recombination deficiency, and altered fraction as well as intratumoral infiltration of T helper type 1 (Th1) and Th2 cells, but lower interferon-gamma response and M2 macrophage infiltration. Cell proliferation-related gene sets in the Hallmark collection (E2F targets, G2M checkpoint, Myc target v1 and mitotic spindle), MTORC1 signaling, DNA repair, PI3K MTOR signaling, and unfolded protein response were all enriched in the MKi67-high HCC (false discovery rate (FDR) < 0.25).
Conclusions: High MKi67 gene expression identified highly proliferative HCC with aggressive biology involving classical pathways in cell cycle regulation and DNA repair, as well as poor overall oncologic outcomes. This suggests potential for personalized treatment strategies, but validation and refinement of these observations require further research to elucidate the underlying mechanisms and validate therapeutic targeting of these pathways in MKi67-high HCC tumors.
Keywords: MKi67; Gene expression; HCC; Outcomes; Signaling pathways
| Introduction | ▴Top |
Hepatocellular carcinoma (HCC) ranks as the fourth leading cause of cancer-related deaths worldwide and is currently the fastest growing cause of cancer-specific mortality in the United States [1, 2]. HCC frequently arises in individuals with chronic liver disease, commonly due to persistent hepatoviral infection or nonalcoholic fatty liver disease (NAFLD) [3, 4]. The prognosis of patients with HCC is generally poor with an overall 5-year survival of approximately 18% [5]. This is because most patients (60%) are diagnosed with an advanced disease, which makes them ineligible for curative intent treatment [6]. Consequently, the identification of precise prognostic biomarkers holds the potential to enhance patient selection and identify those who are more likely to benefit from aggressive HCC treatment.
Ki67 is a protein predominantly found in the nucleolar cortex and exhibits high expression in the majority of proliferating malignant cells, while being seldom expressed in normal cells. Ki67 is recruited into chromosomes during cell division, and its concentration rises during the transition from grade 1 (G1) to mitosis with a rapid decrease in later phases [7-9]. As a result, it is one of the most employed clinical markers for cell proliferation in many malignancies [10-14]. Prior studies have shown that Ki67 expression is correlated with worse tumor biology and poorer overall outcomes in patients with central nervous system malignancies, renal cell carcinoma, adrenocortical carcinoma, and uterine cancer [15, 16]. In patients with HCC, a recent meta-analysis showed that high Ki67 protein expression was associated with larger tumor size, higher number of lymph node metastases, cirrhosis, vascular invasion, and presence of distant metastasis [17].
The most widely adopted method by which Ki67 is assessed and reported is through immunostaining. However, conventional immunostaining is subject to user variability and subjective interpretation, and results are histology-specific [18-20]. In this context, the expression of the Ki67 gene (MKi67), which is more objective and accurately quantifiable than immunohistochemistry, can be employed to identify highly proliferative HCC [21]. We therefore hypothesized that HCC with high MKi67 expression correlates with overall oncologic outcomes, and we sought to investigate its association with aggressive tumor biology.
| Materials and Methods | ▴Top |
Clinical and transcriptomic data acquisition
We utilized The Cancer Genome Atlas (TCGA) that collected the untreated bulk tumors and their associated transcriptomes with the clinicopathological data of each patient as we previously described [22-36]. TCGA is a comprehensive, collaborative initiative that systematically characterizes gene expression profiles across various cancer types. It provides a vast repository of genomic and clinical data, facilitating in-depth analyses to better understand the molecular basis of cancer. There are 358 HCC patients included in TCGA cohort that we used in our previous studies [37-43]. It is important to note, however, that the TCGA dataset lacks certain demographic and clinical characteristics such as patients’ medical history or treatment modalities. Similarly, factors such as smoking, socioeconomic status or diet were inaccessible to the authors.
This dataset was further validated using an additional 115 patients from the Gene Expression Omnibus (GEO) GSE76427 cohort [44]. Tumor characteristics: grade, size, node and staged were obtained from the Genomic Data Commons (GDC) Data Portal and reported according to the American Joint Committee on Cancer (AJCC) classification. We compared the GSE6764 (n = 75) cohort to the GSE89377 (n = 107) cohort [45, 46] to investigate the association between MKi67 expression, clinicopathological characteristics and outcomes from GEO repository. HCC pathological classification in GSE6764 followed the guidelines of the International Working Party and was defined as: 1) Very early HCC (n = 8), well-differentiated tumors < 2 cm in diameter with no vascular invasion/satellites (size range: 8 - 20 mm); 2) Early HCC (n = 10), tumors measuring < 2 cm with microscopic vascular invasion/satellites; well to moderately differentiated tumors measuring 2 - 5 cm without vascular invasion/satellites; or 2 - 3 well-differentiated nodules measuring < 3 cm (size range: 3 - 45 mm); 3) Advanced HCC (n = 7), poorly differentiated tumors measuring > 2 cm with microvascular invasion/satellites or tumors measuring > 5 cm; and 4) Very advanced HCC (n = 10), tumors with macrovascular invasion or diffuse liver involvement. For the GSE89377 cohort (n = 107), they were defined as: normal (n = 13), dysplasia (n = 22), cirrhosis (n = 12), low-grade chronic hepatitis (n = 8), high-grade chronic hepatitis (n = 12), early HCC (n = 5), HCC grade 1 (n = 9), HCC grade 2 (n = 12) and HCC grade 3 (n = 14) [47].
Patients were divided into two groups based on their MKi67 gene expression levels. Those whose MKi67 gene expression levels exceeded the median were designated as the highly proliferative group (MKi67-high), whereas the remaining patients in each cohort were grouped as the MKi67-low group.
All genomic analyses used were log2 transformed normalized transcriptomic data. The TCGA and all GEO cohorts used in this study are deidentified and available within the public domain, therefore the Institutional Review Board was waived. The declaration of ethical compliance with human study was not applicable.
Tumor microenvironment
The xCell algorithm obtained through the xCell website [48], was used to calculate the immune cell infiltration in the tumor microenvironment through transcriptomic data as we described previously [43, 49-51]. The scoring of homologous recombination, intratumor heterogeneity, fraction altered, silent mutation, non-silent mutation, single-nucleotide variant (SNV) neoantigens, indel neoantigens, leukocyte fraction, lymphocyte infiltration, and interferon (IFN)- response score was performed as published by Thorsson et al [52].
Gene set enrichment analysis (GSEA)
The publicly-available software (GSEA version 4.0.3) and the GSEA algorithm were used in this study [53]. Statistical significance was determined to have a false discovery rate (FDR) of 0.25.
Statistical analysis
Statistical significance for comparison analysis between groups was set at P less than 0.05 by the Kruskal-Wallis test, the Mann-Whitney U test and two-tail Fisher’s exact tests. Survival analysis was performed with the Kaplan-Meier method and log rank test. Statistical analyses and data plotting were performed in R software version 4.2.3 (R Project for Statistical Computing).
| Results | ▴Top |
MKi67-high HCC patients had worse disease-free, disease-specific, and overall survival (OS) in the TCGA cohort
First, relationships between MKi67 expression and survival outcomes were assessed. MKi67-high expression was significantly associated with worse disease-free survival (DFS), disease-specific survival (DSS) and OS, when compared to MKi67-low expression in TCGA (P < 0.001 for all comparisons), although these results were not validated in in GEO GSE766427 cohort (OS P = 0.364 and PFS P = 0.751) (Fig. 1).
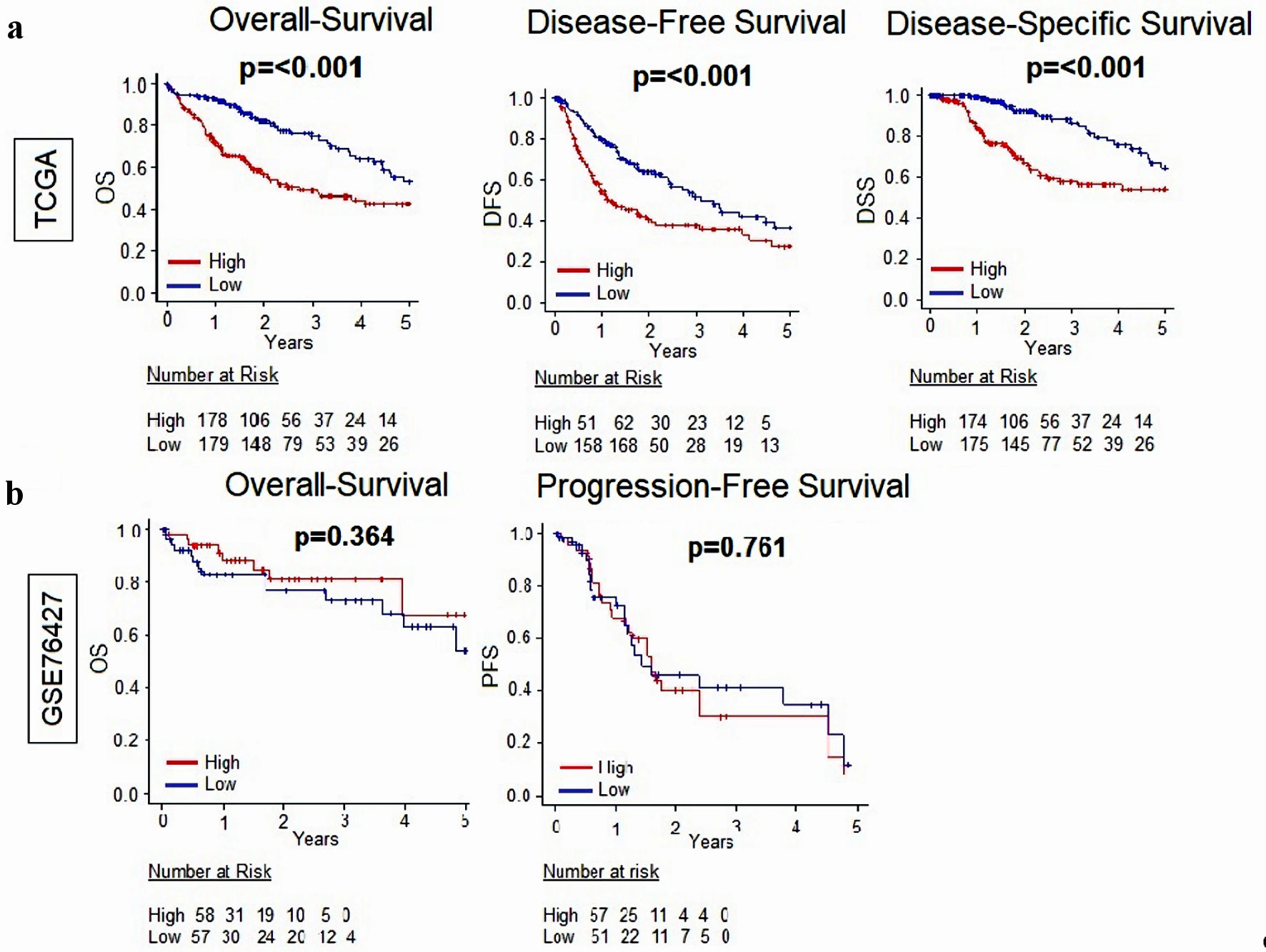 Click for large image | Figure 1. Relationship between MKi67 and survival outcomes in patients with HCC. (a) Kaplan-Meier survival curves comparing high- vs. low-MKi67 expression in HCC to determine disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) in the TCGA (n = 358) cohort. (b) Kaplan-Meier survival curves comparing high- vs. low-MKi67 expression in HCC to determine overall survival and progression-free survival in the GSE76427 (n = 115) cohort. The P value was calculated using a log rank test. Significant P value < 0.05. HCC: hepatocellular carcinoma; TCGA: The Cancer Genome Atlas. |
Given that higher histological grade is a pathological determination of cancer cell proliferation, we did a subgroup analysis of MKi67-high vs. low stratified by histological grade (Fig. 2). Our results demonstrated that MKi67-high had worse oncologic outcomes across all histological grades, but it is more pronounced in histological grade 3 (G3) tumors. Nevertheless, DFS in G1 and OS in grade 2 (G2) tumors did not reach statistical significance (P = 0.978 and P = 0.072, respectively).
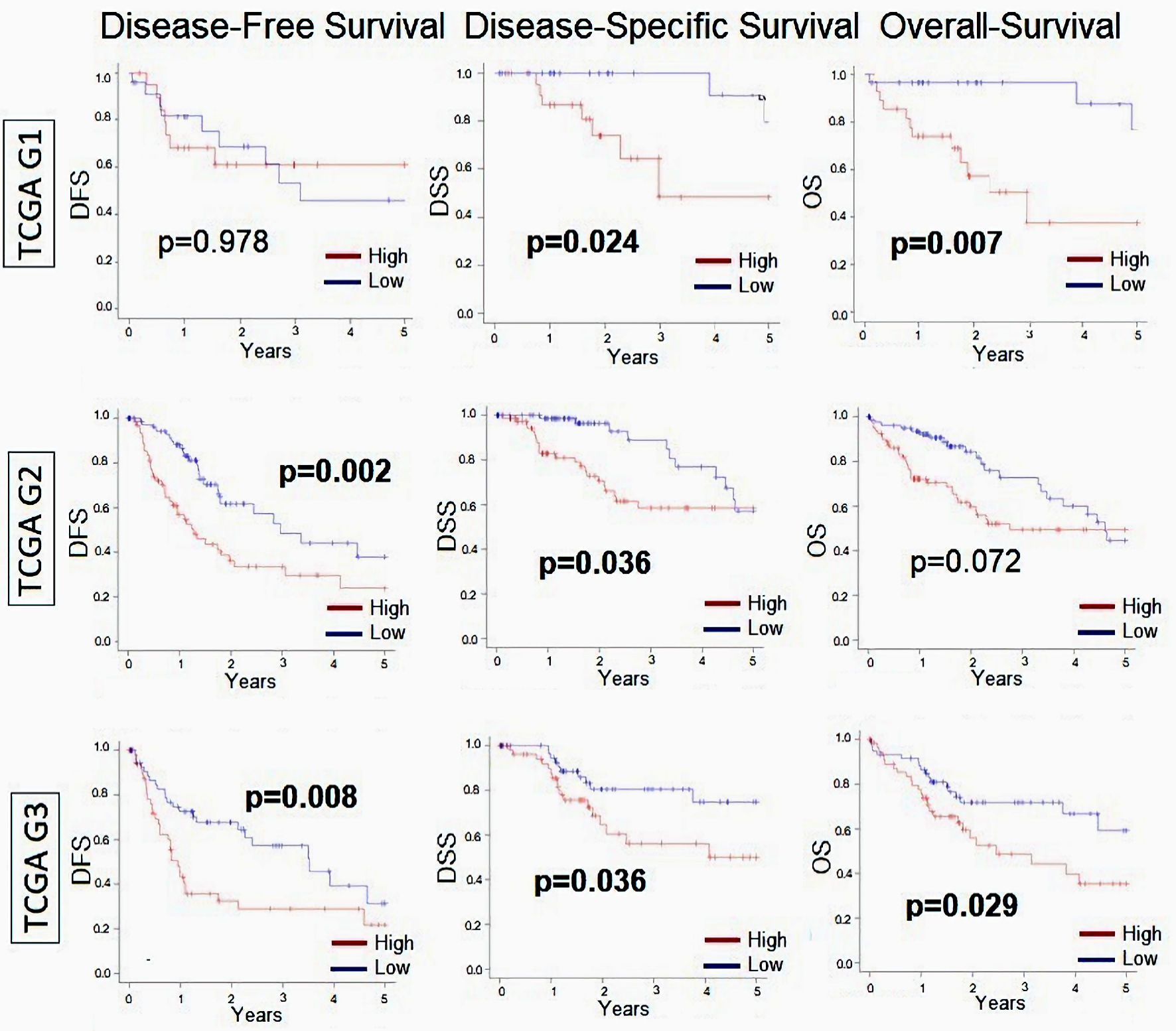 Click for large image | Figure 2. Relationship between MKi67 and survival outcomes in patients with HCC broken down by histological grade. Kaplan-Meier survival curves comparing high- vs. low-MKi67 expression in HCC to determine disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) in grade 1 (n = 53), grade 2 (n = 168), and grade 3 (n = 121) from TCGA cohort. The P value was calculated using a log rank test. Significant P value < 0.05. HCC: hepatocellular carcinoma; TCGA: The Cancer Genome Atlas. |
MKi67 expression was positively correlated with HCC progression
Given that MKi67 was associated with worse oncologic outcomes, we hypothesized that MKi67 expression increases in a stepwise progression from normal liver to early HCC to advanced HCC. MKi67 expression was therefore measured at each phase of histological progression in the GSE6764 and GSE89377 cohorts (Fig. 3). We found that MKi67 expression increased in a stepwise fashion from early to advanced HCC (P ≤ 0.001) with no significant difference between the normal, dysplastic, and cirrhotic liver in the GSE6764 cohort (normal liver vs. early HCC P = 0.015; normal vs. advanced HCC P < 0.001; normal vs. very advanced HCC, P < 0.001). We validated these results with the GSE89377 cohort, showing MKi67 expression was also significantly enhanced in higher grades of HCC compared with normal, dysplastic, and cirrhotic liver and low- and high-grade chronic hepatitis (normal vs. G1, P = 0.025; normal vs. G2, P = 0.025; normal vs. G3, P < 0.001).
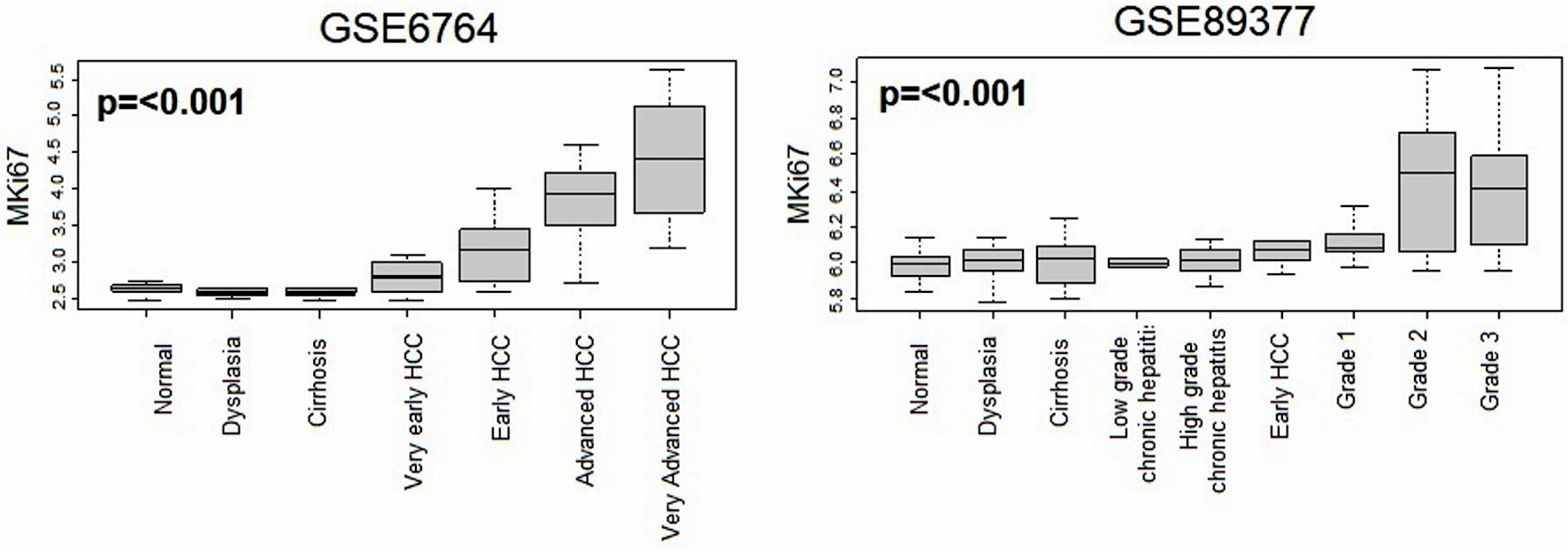 Click for large image | Figure 3. Relationship between MKi67 expression and histological progression of HCC. Boxplots of the MKi67 expression by multistep hepatocarcinogenesis, including normal liver tissue (n = 10), dysplasia (n = 17), cirrhosis (n = 13), very early HCC (n = 8), early HCC (n = 10), advanced HCC (n = 7), and very advanced HCC (n = 10) in the GSE6764 (n = 75), The P value was calculated using a Kruskal-Wallis test. HCC: hepatocellular carcinoma. |
MKi67 expression correlated with histological grade, tumor size, lymph node metastasis and AJCC stage in HCC
Next, we investigated whether MKi67 expression was associated with aggressive tumor characteristics in HCC. To this end, we evaluated the differences in MKi67 gene expression stratified by histological grade, T-category, N-category, and staging by AJCC as a clinical marker of tumor aggressiveness. Child-Pugh classification was also assessed as a marker of severity of liver cirrhosis. As expected, MKi67 expression strongly correlated with histological grade (Fig. 4) (P ≤ 0.001). We found that MKi67 expression correlated with a higher T-stage (P < 0.001), although it was less clear in T4 tumors noting the small sample size. Node-positive tumors showed a non-significant trend toward higher expression of MKi67 (P = 0.053). MKi67 expression significantly correlated with higher stage consistently in both TCGA and GSE76427cohorts (P ≤ 0.001), with the exception of metastatic (stage IV) tumors. In terms of cirrhosis, normal liver showed the highest MKi67 expression. Notably, there was no statistically significant difference in MKi67 expression among any Child-Pugh class in the GSE76427 cohort (P = 0.208).
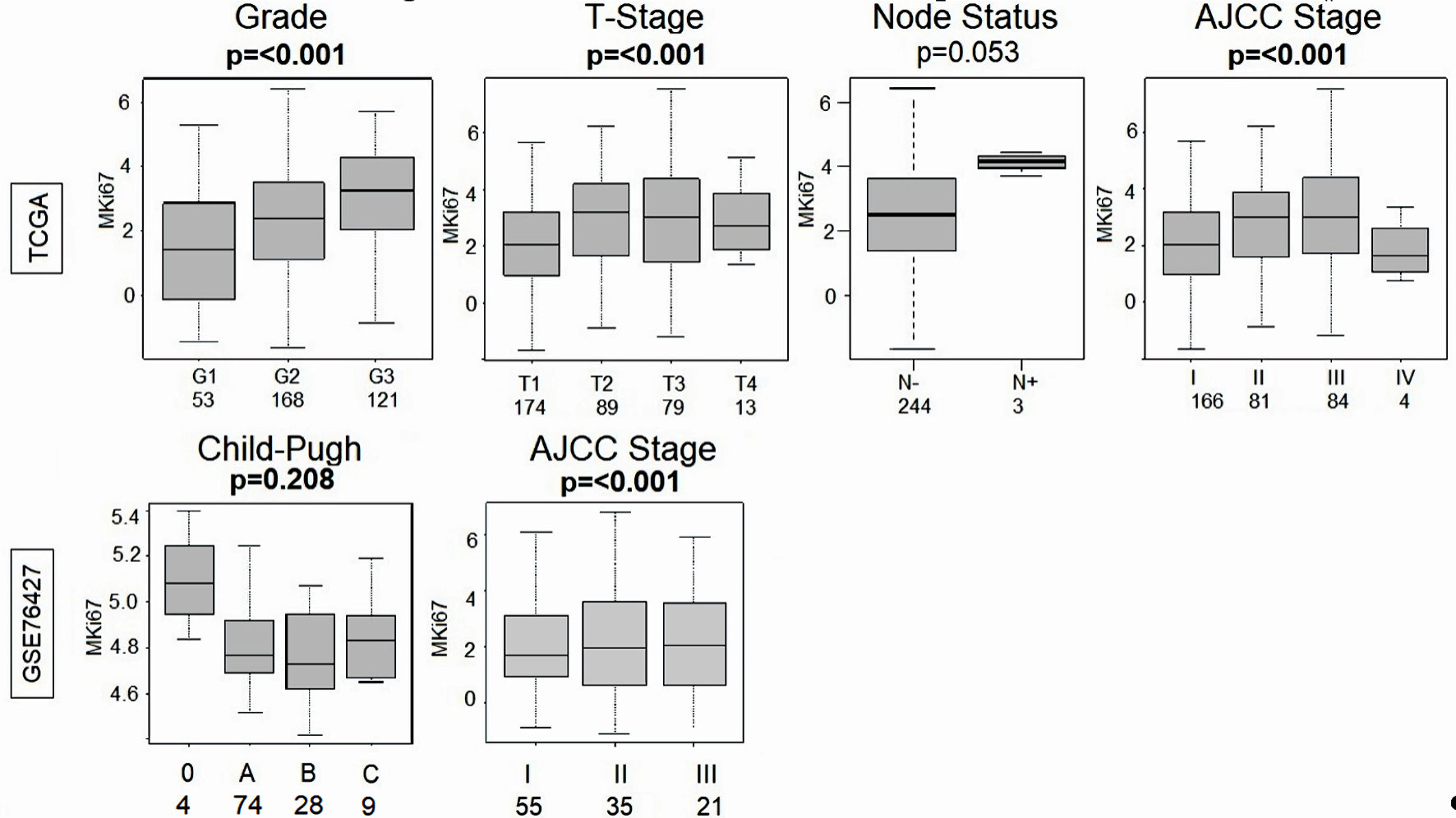 Click for large image | Figure 4. Comparison of the MKi67 expression and grade, T-stage, node status and AJCC stage in the TCGA (n = 358) cohort and MKi67 expression and Child-Pugh classification and AJCC stage in the GSE76427 (n = 115) cohort. P value < 0.05 was considered statistically significant. TCGA: The Cancer Genome Atlas; AJCC: American Joint Committee on Cancer. |
MKi67-high was significantly associated with intratumor heterogeneity, homologous recombination defects, and altered fraction
To understand the genomic profile in HCC with high versus low expression of MKi67, we evaluated the underlying mutation rate of HCC using scores pre-calculated by Thorsson et al [52]. We found that homologous recombination defects, intratumor heterogeneity and altered fraction were significantly higher in the MKi67-high HCC (P ≤ 0.001), but not silent mutation rate (P = 0.601), non-silent mutation rate (P = 0.830), SNV neoantigens (P = 0.438), or indel neoantigens (P = 0.745) in the TCGA cohort (Fig. 5).
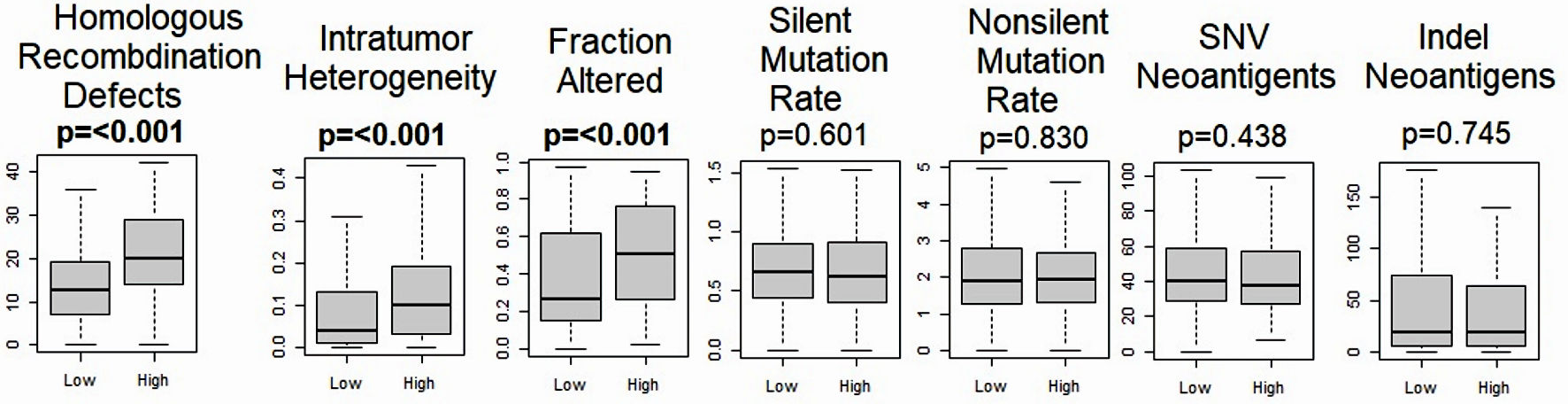 Click for large image | Figure 5. Relationship between MKi67 and mutation related scores. Boxplots of the comparison of the high- vs. low-MKi67 expression and homologous recombination deficiency, intratumor heterogeneity, fraction altered, Silent and non-silent mutation rates, single-nucleotide variant (SNV) neoantigens, and indel neoantigens. The P value was calculated using the Mann-Whitney U test. Significant P value < 0.05. |
MKi67-high HCC was not consistently associated with immune cell infiltrations, except for T helper type 1 (Th1) and Th2 cells
In view of the higher homologous recombination defects but not mutation rates in MKi67-high HCC tumors, we sought to evaluate the relationship between MKi67 expression and immune cell infiltration in the tumor microenvironment. Overall, there was no association between MKi67 expression and lymphocyte infiltration, leukocyte fraction, or transforming growth factor (TGF)-beta response and (Fig. 6a). Interestingly, IFN-gamma response was attenuated in MKi67-high tumors (P = 0.002). The only immune cells that were highly represented in MKi67-high tumor microenvironment were Th1 and Th2 cells. In contrast, M2 macrophages were significantly overrepresented in the MKi67-low tumor microenvironment. Moreover, while alpha-fetoprotein (AFP) exhibited a significant elevation in MKi67-high tumors within the TCGA cohort, this association did not reach statistical significance in the GSE76427 cohort (Fig. 6b).
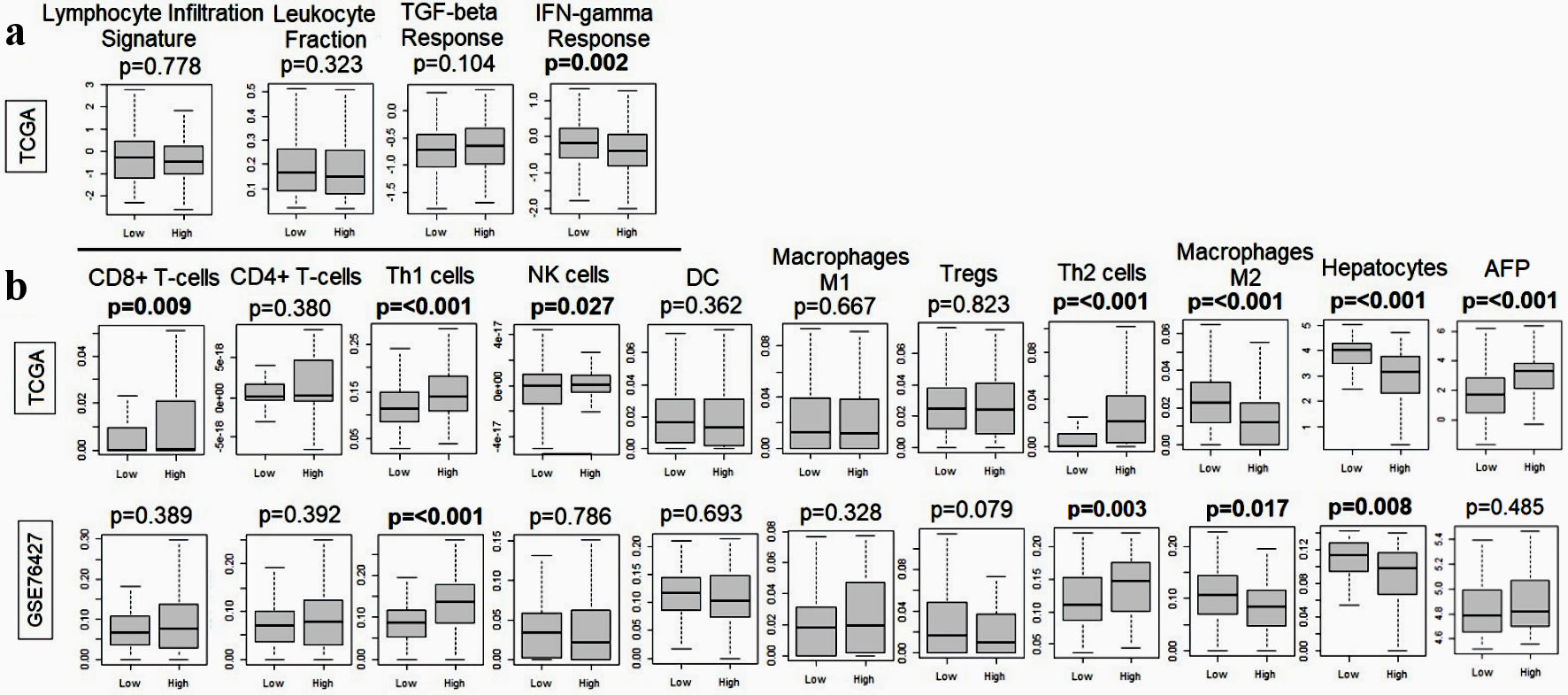 Click for large image | Figure 6. Relationship between MKi67 and immune response-related genes in the TCGA and GSE76427. (a) Boxplots of lymphocyte infiltration signature, leukocyte fraction, TGF-beta response, and IFN-gamma response. (b) Boxplots of anticancer immune cells: CD8+ T cell, CD4+ T cell, T helper type 1 (Th1) cells, M1 macrophages and dendritic cells and pro-cancer immune cells including regulatory T cells (Treg), T helper type 2 (Th2) cells, M2 macrophages, hepatocytes and alpha-fetoprotein (AFP). The P value was calculated using the Mann-Whitney U test. Significant P value < 0.05. TGF: transforming growth factor; IFN: interferon; TCGA: The Cancer Genome Atlas; NK: natural killer; DC: dendritic cell. |
MKi67-high HCC is associated with cell proliferation-related and cell-cycle gene sets
Considering MKi67-high HCC was associated with aggressive clinical characteristics, we conducted a GSEA in the TCGA and GSE76427 cohorts to investigate enriched pathways associated with MKi67-high tumors. As expected, we found significant enrichment of cell proliferation-related gene sets in the Hallmark collection: E2F targets, mitotic spindle, G2M checkpoints, Myc targets V1 (Fig. 7). Furthermore, we found that gene sets that reflect aggressive tumor biology: unfolded protein response (UPR), PI3K MTOR signaling, MTORC1 signaling and DNA repair, were all enriched to MKi67-high HCC in both TCGA and GSE76427 cohorts.
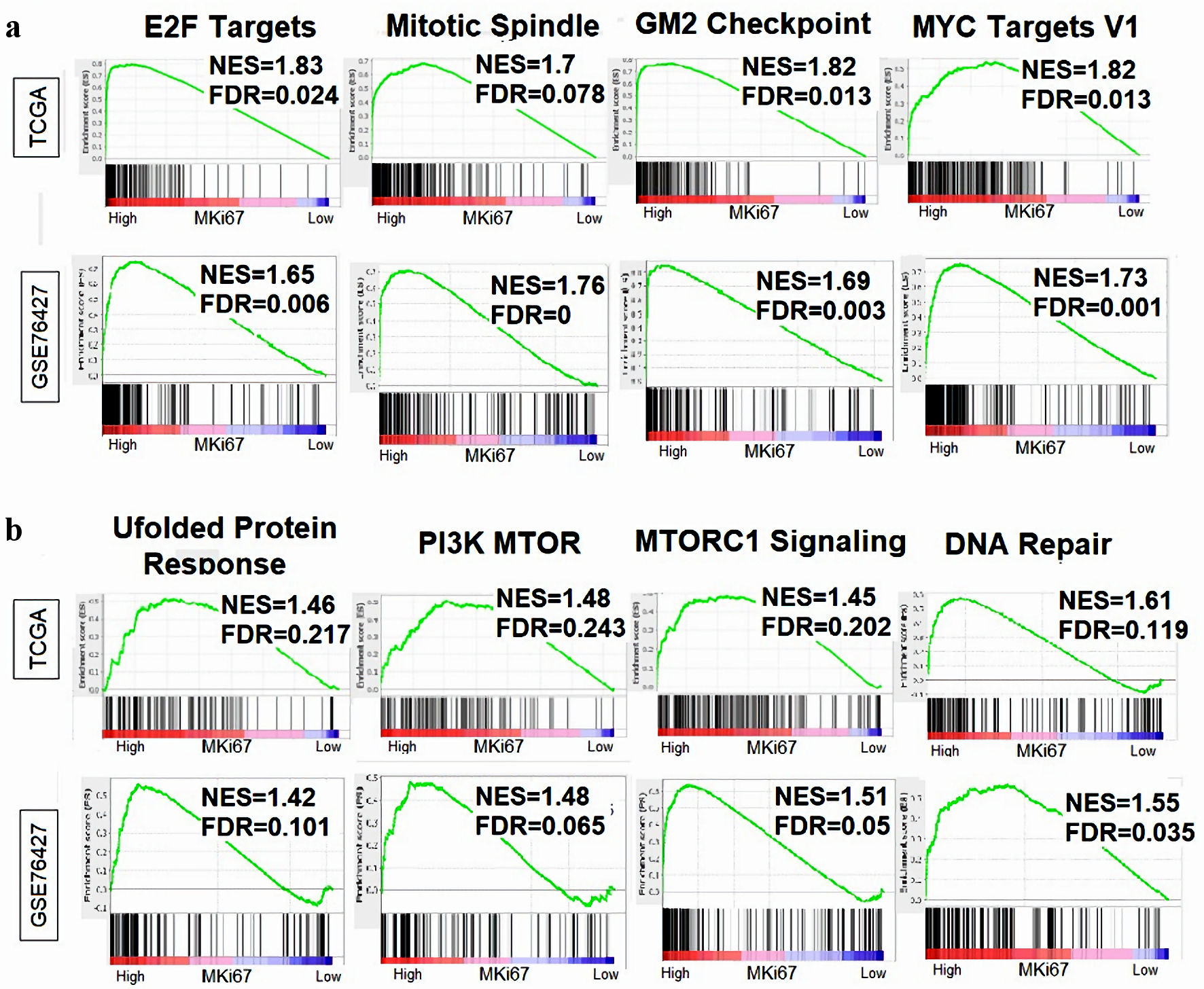 Click for large image | Figure 7. Relationship between MKi67 expression and cell proliferation-related gene sets. Gene Set Enrichment Analysis (GSEA) was performed on the Hallmark gene sets, comparing MKi67-high vs. low scores in HCC across the TCGA and GSE76427 cohorts Enrichment plots was generated displaying the normalized enrichment score (NES) and false discovery rate (FDR) for proliferation-related gene sets. An FDR of 0.25 was determined statistically significant as recommended by the GSEA software. (a) Cell proliferation-related gene sets in the Hallmark collection. (b) Genes related to aggressive tumor biology. HCC: hepatocellular carcinoma; TCGA: The Cancer Genome Atlas. |
| Discussion | ▴Top |
In this study, we found that high expression of MKi67 was associated with biologically aggressive HCC. Not only was higher expression of MKi67 positively correlated with aggressive clinical tumor characteristics (histological grade, tumor size, and AJCC staging), but also intratumor heterogeneity, homologous recombination defects, and altered fraction. Further, in our analysis of the GSE6764 and GSE89377 cohorts, MKi67 expression appeared to correlate with progression of HCC from early to advanced and histological grade from lower (G1) to higher (G2 and G3) in a stepwise fashion. Interestingly, despite higher cell proliferation in MKi67-high HCC, there was no association with the degree of immune response (except for increased infiltrating Th1 and Th2 cells). More importantly, high expression of MKi67 in HCC was associated with enriched expression of multiple genes involved in the cell cycle and DNA repair pathways.
Tumor markers play a crucial role in the diagnosis and prognosis of various cancers. In the context of HCC, AFP is the most extensively utilized biomarker for both diagnosis and prognostication [54]. Our study demonstrated a positive correlation of AFP levels and MKi67 expression in the TCGA cohort but not in the GSE76427. While not directly connected to Ki67, AFP exhibits a dual regulatory role in cell proliferation. The impact of AFP on growth regulation, whether enhancing or inhibitory, is contingent upon the concentration of AFP and the levels of cytokines, hormones, and growth factors in the culture system [55]. However, it is worth noting that approximately 30-40% of HCC cases are AFP-negative. While a lack of detectable AFP in the serum may be indicative of positive outcomes in liver cancer cases, it can also lead to an inaccurate diagnosis of liver cancer as a whole [56].
Ki67 expression, on the other hand, serves as a comprehensive surrogate measure of cell proliferation, and heightened expression is indicative of aggressive tumor behavior. Numerous studies support the pivotal role of Ki67 in cancer prognostics, as its expression is strongly correlated with the aggressiveness of various tumors, including those affecting the breast, pancreas, lungs, central nervous system, prostate, and salivary glands [57-63]. In resonance with this study, Luo et al found that high Ki67 protein expression was associated with more advanced HCC stages, poorer differentiation, larger tumors, as well as poorer DFS, RFS and OS [17]. However, in our study, Ki67 expression was evaluated by directly examining MKi67 gene expression. This approach has been proven to be more accurate in gauging tumor proliferation compared to assessing protein expression through immunohistochemistry (IHC). This superiority is attributed to its higher sensitivity, reproducibility, and reduced variability among users [64].
We found that intratumor heterogeneity, homologous recombination defects and altered fraction were significantly higher in the MKi67-high HCC. Intratumor heterogeneity has been found to influence tumor growth, metastasis, recurrence, and resistance to cytotoxic chemotherapy [65-68]. Similarly, homologous recombination defects have been associated with genomic scarring and a poor prognosis in HCC [69]. Additionally, as reported by Liu et al, HCC tumors with higher altered fraction are associated with rapid proliferation and immune evasion and could result in an attenuated response to immunotherapy [70, 71].
This study revealed no association between lymphocyte infiltration, leukocyte fraction or TGF-beta response and MKi67 expression. Additionally, there was no significant increase in pro-cancer immune cell proliferation except for Th1 and Th2 cells in the MKi67-high tumors. A plausible explanation for this weakened anticancer immune response might involve T cell exhaustion, a phenomenon previously explored by Wu et al [72]. Despite the association of MKi67 upregulation with increased infiltration of B cells, CD4+ T cells, CD8+ T cells, neutrophils, dendritic cells, and other functional T cells, MKi67 may concurrently contribute to T cell exhaustion. This dual effect could result in a diminished anticancer immune response, characterized by the upregulation of critical genes such as TIM-3 and TIGIT. These genes, comparable to the therapeutic targets programmed death ligand-1 (PD-1) and cytotoxic T lymphocyte antigen (CTLA), play a role in regulating T-cell responses and could act as therapeutic targets for immunotherapy drugs theoretically with fewer toxicities due to their more targeted activity [72].
To validate the association between pathological and biological characteristics with MKi67 expression, we conducted a gene-set enrichment analysis. We found that in both TCGA and the GSE76427 cohorts, MKi67-high HCC had significant enrichment of E2F targets, mitotic spindle, G2M checkpoints, Myc targets V1, UPR, PI3K MTOR signaling, MTORC1 signaling, and DNA repair pathways. These cell proliferation-related and cell-cycle gene pathways genes have an important role in the pathogenesis of HCC. Our previous study showed that upregulation of individual E2F targets was associated with worse OS and DFS in HCC, especially the alterations in E2F3, E2F5 and E2F6 [73]. Furthermore, a metanalysis of c-Myc overexpression in HCC was associated with worse oncologic outcomes [74]. Even though the PI3K/Akt/mTOR pathway is upregulated in approximately 50% of HCC patients and plays an important role in the pathogenesis of this malignancy, clinical studies have failed to demonstrate any capacity of mTOR inhibitors to decrease tumor growth or recurrence [75, 76]. Moreover, our group’s prior study on the role of the UPR in HCC demonstrated that the upregulation of UPR was associated with multiple parameters of cell proliferation, including MKi67 expression, enrichment of cell proliferation-related gene sets and mutational load that ultimately translated to worse survival of patients with HCC [39]. Similarly, Oshi et al showed that the DNA repair pathway was enhanced in higher histological grade HCCs, which also burdened with elevated tumor heterogeneity and higher mutational load ultimately yielding worse survival outcomes compared to HCCs without enriched DNA repair pathway [77].
This study provides evidence supporting the potential use of MKi67 expression as a prognostic marker in HCC and sheds light on its underlying biological mechanisms. These findings suggest that HCC tumors with high MKi67 expression may exhibit unique molecular characteristics. These characteristics, particularly those related to DNA repair pathways and immune responses, could potentially guide treatment strategies. For instance, we speculate that HCC with high MKi67 expression might be more responsive to therapies targeting cell cycle regulation, DNA repair pathways, or immunomodulation. We highlight the possibility that these findings could inform personalized treatment strategies, with further research needed to validate and refine these speculations.
There are some limitations to our study. As we used publicly available cohorts, our results may not reflect the heterogeneity among the patients in the population. Unfortunately, our group did not have access to information regarding the patients’ medical history or treatment modalities such as surgery, local ablative, loco-regional or systemic therapy to the cohorts we analyzed. Additionally, the absence of detailed clinical follow-up data in some cases limits the ability to draw robust conclusions about responses to treatments. Similarly, factors such as smoking, socioeconomic status or diet were inaccessible to the authors.
In conclusion, our study provides compelling evidence that high expression of MKi67 in HCC is closely linked to a more aggressive biological phenotype driven by the upregulation of multiple interconnected pathways involved in cell cycle regulation, DNA repair and oncogenic signaling. Further clinical studies are warranted to elucidate the underlying mechanisms and validate the therapeutic potential of targeting these pathways in MKi67-high HCC tumors. Such efforts hold the promise of improving patient outcomes and advancing precision medicine approaches for the management of HCC.
Acknowledgments
K. Takabe was supported by US National Institutes of Health grants R37CA248018, R01CA250412, R01CA251545, R01EB029596, as well as US Department of Defense BCRP grants W81XWH-19-1-0674 and W81XWH-19-1-0111. This work was also supported by Roswell Park Comprehensive Cancer Center and National Cancer Institute cancer center support grant P30CA016056.
Financial Disclosure
None to declare.
Conflict of Interest
The authors have no potential conflicts of interest to disclose.
Informed Consent
Not applicable.
Author Contributions
Conceptualization: V. Ramos-Santillan, M. Oshi, and K. Takabe. Data analyses: M. Oshi, I. Endo, and K. Takabe. Writing - original draft preparation: V. Ramos-Santillan. Writing - review and editing: E. Nelson, M. Oshi, I. Endo, and K. Takabe. Supervision: I. Endo, and K. Takabe. Funding acquisition: K. Takabe. All authors have read and agreed to the published version of the manuscript.
Data Availability
The data that support the findings of this study are available from the corresponding author, KT, upon reasonable request.
Abbreviations
HCC: hepatocellular carcinoma; TME: tumor microenvironment; TCGA: The Cancer Genome Atlas; GEO: Gene Expression Omnibus; DFS: disease-free survival; DSS: disease-specific survival; OS: overall survival; NAFLD: nonalcoholic fatty liver disease; GDC: Genomic Data Commons; AJCC: American Joint Committee on Cancer; SNV: single-nucleotide variant; IFN: interferon; GSEA: Gene enrichment analysis; G1: grade1; G2: grade 2; G3: grade 3
| References | ▴Top |
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17-48.
doi pubmed - Kanwal F, Singal AG. Surveillance for hepatocellular carcinoma: current best practice and future direction. Gastroenterology. 2019;157(1):54-64.
doi pubmed pmc - Ganne-Carrie N, Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol. 2019;70(2):284-293.
doi pubmed - Mahmud N, Fricker Z, Hubbard RA, Ioannou GN, Lewis JD, Taddei TH, Rothstein KD, et al. Risk Prediction Models for Post-Operative Mortality in Patients With Cirrhosis. Hepatology. 2021;73(1):204-218.
doi pubmed pmc - Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380(15):1450-1462.
doi pubmed - Chapman WC, Klintmalm G, Hemming A, Vachharajani N, Majella Doyle MB, DeMatteo R, Zaydfudim V, et al. Surgical treatment of hepatocellular carcinoma in North America: can hepatic resection still be justified? J Am Coll Surg. 2015;220(4):628-637.
doi pubmed - Isola J, Helin H, Kallioniemi OP. Immunoelectron-microscopic localization of a proliferation-associated antigen Ki-67 in MCF-7 cells. Histochem J. 1990;22(9):498-506.
doi pubmed - Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311-322.
doi pubmed - Verheijen R, Kuijpers HJ, Schlingemann RO, Boehmer AL, van Driel R, Brakenhoff GJ, Ramaekers FC. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. I. Intracellular localization during interphase. J Cell Sci. 1989;92(Pt 1):123-130.
doi pubmed - Karamitopoulou E, Perentes E, Tolnay M, Probst A. Prognostic significance of MIB-1, p53, and bcl-2 immunoreactivity in meningiomas. Hum Pathol. 1998;29(2):140-145.
doi pubmed - Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep. 2015;11(3):1566-1572.
doi pubmed - Zheng K, Zhu W, Tan J, Wu W, Yang S, Zhang J. Retrospective analysis of a large patient sample to determine p53 and Ki67 expressions in renal cell carcinoma. J BUON. 2014;19(2):512-516.
pubmed - Zini L, Porpiglia F, Fassnacht M. Contemporary management of adrenocortical carcinoma. Eur Urol. 2011;60(5):1055-1065.
doi pubmed - Zizi-Sermpetzoglou A, Moustou E, Petrakopoulou N, Arkoumani E, Tepelenis N, Savvaidou V. Atypical polypoid adenomyoma of the uterus. A case report and a review of the literature. Eur J Gynaecol Oncol. 2012;33(1):118-121.
pubmed - Yang C, Su H, Liao X, Han C, Yu T, Zhu G, Wang X, et al. Marker of proliferation Ki-67 expression is associated with transforming growth factor beta 1 and can predict the prognosis of patients with hepatic B virus-related hepatocellular carcinoma. Cancer Manag Res. 2018;10:679-696.
doi pubmed pmc - Yang C, Zhang J, Ding M, Xu K, Li L, Mao L, Zheng J. Ki67 targeted strategies for cancer therapy. Clin Transl Oncol. 2018;20(5):570-575.
doi pubmed - Luo Y, Ren F, Liu Y, Shi Z, Tan Z, Xiong H, Dang Y, et al. Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: a meta-analysis. Int J Clin Exp Med. 2015;8(7):10235-10247.
pubmed pmc - Nadler A, Cukier M, Rowsell C, Kamali S, Feinberg Y, Singh S, Law CH. Ki-67 is a reliable pathological grading marker for neuroendocrine tumors. Virchows Arch. 2013;462(5):501-505.
doi pubmed - Nagao K, Yamamoto Y, Hara T, Komatsu H, Inoue R, Matsuda K, Matsumoto H, et al. Ki67 and BUBR1 may discriminate clinically insignificant prostate cancer in the PSA range <4 ng/ml. Jpn J Clin Oncol. 2011;41(4):555-564.
doi pubmed - Rioux-Leclercq N, Turlin B, Bansard J, Patard J, Manunta A, Moulinoux JP, Guille F, et al. Value of immunohistochemical Ki-67 and p53 determinations as predictive factors of outcome in renal cell carcinoma. Urology. 2000;55(4):501-505.
doi pubmed - Noske A, Loibl S, Darb-Esfahani S, Roller M, Kronenwett R, Muller BM, Steffen J, et al. Comparison of different approaches for assessment of HER2 expression on protein and mRNA level: prediction of chemotherapy response in the neoadjuvant GeparTrio trial (NCT00544765). Breast Cancer Res Treat. 2011;126(1):109-117.
doi pubmed - Asaoka M, Patnaik SK, Zhang F, Ishikawa T, Takabe K. Lymphovascular invasion in breast cancer is associated with gene expression signatures of cell proliferation but not lymphangiogenesis or immune response. Breast Cancer Res Treat. 2020;181(2):309-322.
doi pubmed pmc - Gandhi S, Elkhanany A, Oshi M, Dai T, Opyrchal M, Mohammadpour H, Repasky EA, et al. Contribution of immune cells to glucocorticoid receptor expression in breast cancer. Int J Mol Sci. 2020;21(13):4635.
doi pubmed pmc - Narayanan S, Kawaguchi T, Peng X, Qi Q, Liu S, Yan L, Takabe K. Tumor infiltrating lymphocytes and macrophages improve survival in microsatellite unstable colorectal cancer. Sci Rep. 2019;9(1):13455.
doi pubmed pmc - Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, et al. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20(11):2655.
doi pubmed pmc - Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, et al. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20(17):4197.
doi pubmed pmc - Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, et al. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21(18):6968.
doi pubmed pmc - Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, et al. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel). 2020;12(5):1148.
doi pubmed pmc - Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, et al. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21(18):6708.
doi pubmed pmc - Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. Inflammation is associated with worse outcome in the whole cohort but with better outcome in triple-negative subtype of breast cancer patients. J Immunol Res. 2020;2020:5618786.
doi pubmed pmc - Oshi M, Tokumaru Y, Patel A, Yan L, Matsuyama R, Endo I, Katz MHG, et al. A novel four-gene score to predict pathologically complete (r0) resection and survival in pancreatic cancer. Cancers (Basel). 2020;12(12):3635.
doi pubmed pmc - Ramanathan R, Olex AL, Dozmorov M, Bear HD, Fernandez LJ, Takabe K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat. 2017;162(1):191-198.
doi pubmed pmc - Takahashi H, Katsuta E, Yan L, Tokumaru Y, Katz MHG, Takabe K. Transcriptomic profile of lymphovascular invasion, a known risk factor of pancreatic ductal adenocarcinoma metastasis. Cancers (Basel). 2020;12(8):2033.
doi pubmed pmc - Takeshita T, Torigoe T, Yan L, Huang JL, Yamashita H, Takabe K. The impact of immunofunctional phenotyping on the malfunction of the cancer immunity cycle in breast cancer. Cancers (Basel). 2020;13(1):110.
doi pubmed pmc - Takeshita T, Yan L, Peng X, Kimbung S, Hatschek T, Hedenfalk IA, Rashid OM, et al. Transcriptomic and functional pathway features were associated with survival after pathological complete response to neoadjuvant chemotherapy in breast cancer. Am J Cancer Res. 2020;10(8):2555-2569.
pubmed pmc - Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, et al. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10(3):897-907.
pubmed pmc - Takahashi H, Kawaguchi T, Yan L, Peng X, Qi Q, Morris LGT, Chan TA, et al. Immune cytolytic activity for comprehensive understanding of immune landscape in hepatocellular carcinoma. Cancers (Basel). 2020;12(5):1221.
doi pubmed pmc - Satyananda V, Oshi M, Tokumaru Y, Maiti A, Hait N, Matsuyama R, Endo I, et al. Sphingosine 1-phosphate (S1P) produced by sphingosine kinase 1 (SphK1) and exported via ABCC1 is related to hepatocellular carcinoma (HCC) progression. Am J Cancer Res. 2021;11(9):4394-4407.
pubmed pmc - Patel A, Oshi M, Yan L, Matsuyama R, Endo I, Takabe K. The unfolded protein response is associated with cancer proliferation and worse survival in hepatocellular carcinoma. Cancers (Basel). 2021;13(17):4443.
doi pubmed pmc - Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, et al. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in hepatocellular carcinoma (HCC). Cancers (Basel). 2021;13(2):323.
doi pubmed pmc - Mukhopadhyay S, Tokumaru Y, Oshi M, Endo I, Yoshida K, Takabe K. Low adipocyte hepatocellular carcinoma is associated with aggressive cancer biology and with worse survival. Am J Cancer Res. 2022;12(8):4028-4039.
pubmed pmc - Hoki T, Katsuta E, Yan L, Takabe K, Ito F. Low DMT1 expression associates with increased oxidative phosphorylation and early recurrence in hepatocellular carcinoma. J Surg Res. 2019;234:343-352.
doi pubmed pmc - Cherkassky L, Oshi M, Abdelfatah E, Wu R, Takabe Y, Yan L, Endo I, et al. An immune-inflamed tumor microenvironment as defined by CD8 score is associated with favorable oncologic outcomes in hepatocellular carcinoma independent of measures of tumor mutational burden. Am J Cancer Res. 2022;12(7):3099-3110.
pubmed pmc - Grinchuk OV, Yenamandra SP, Iyer R, Singh M, Lee HK, Lim KH, Chow PK, et al. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol Oncol. 2018;12(1):89-113.
doi pubmed pmc - Identifying novel drivers of human hepatocellular carcinoma and revealing clinical relevance as early diagnostic and prognostic biomarker, geo, V1. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89377.
- Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938-947.
doi pubmed - International Working Party. Terminology of nodular hepatocellular lesions. Hepatology. 1995;22(3):983-993.
doi pubmed - Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
doi pubmed pmc - Oshi M, Gandhi S, Wu R, Asaoka M, Yan L, Yamada A, Yamamoto S, et al. Development of a novel BRCAness score that predicts response to PARP inhibitors. Biomark Res. 2022;10(1):80.
doi pubmed pmc - Oshi M, Patel A, Wu R, Le L, Tokumaru Y, Yamada A, Yan L, et al. Enhanced immune response outperform aggressive cancer biology and is associated with better survival in triple-negative breast cancer. NPJ Breast Cancer. 2022;8(1):92.
doi pubmed pmc - Oshi M, Gandhi S, Yan L, Tokumaru Y, Wu R, Yamada A, Matsuyama R, et al. Abundance of reactive oxygen species (ROS) is associated with tumor aggressiveness, immune response, and worse survival in breast cancer. Breast Cancer Res Treat. 2022;194(2):231-241.
doi pubmed pmc - Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, et al. The immune landscape of cancer. Immunity. 2019;51(2):411-412.
doi pubmed - Bild A, Febbo PG. Application of a priori established gene sets to discover biologically important differential expression in microarray data. Proc Natl Acad Sci U S A. 2005;102(43):15278-15279.
doi pubmed pmc - Fu CC, Wei CY, Chu CJ, Lee PC, Huo TI, Huang YH, Chao Y, et al. The outcomes and prognostic factors of patients with hepatocellular carcinoma and normal serum alpha fetoprotein levels. J Formos Med Assoc. 2023;122(7):593-602.
doi pubmed - Chen T, Dai X, Dai J, Ding C, Zhang Z, Lin Z, Hu J, et al. AFP promotes HCC progression by suppressing the HuR-mediated Fas/FADD apoptotic pathway. Cell Death Dis. 2020;11(10):822.
doi pubmed pmc - Chi X, Jiang L, Yuan Y, Huang X, Yang X, Hochwald S, Liu J, et al. A comparison of clinical pathologic characteristics between alpha-fetoprotein negative and positive hepatocellular carcinoma patients from Eastern and Southern China. BMC Gastroenterol. 2022;22(1):202.
doi pubmed pmc - Chiloiro S, Bianchi A, Doglietto F, de Waure C, Giampietro A, Fusco A, Iacovazzo D, et al. Radically resected pituitary adenomas: prognostic role of Ki 67 labeling index in a monocentric retrospective series and literature review. Pituitary. 2014;17(3):267-276.
doi pubmed - Jakobsen JN, Sorensen JB. Clinical impact of ki-67 labeling index in non-small cell lung cancer. Lung Cancer. 2013;79(1):1-7.
doi pubmed - Jamali M, Chetty R. Predicting prognosis in gastroentero-pancreatic neuroendocrine tumors: an overview and the value of Ki-67 immunostaining. Endocr Pathol. 2008;19(4):282-288.
doi pubmed - Johannessen AL, Torp SH. The clinical value of Ki-67/MIB-1 labeling index in human astrocytomas. Pathol Oncol Res. 2006;12(3):143-147.
doi pubmed - Moul JW. Angiogenesis, p53, bcl-2 and Ki-67 in the progression of prostate cancer after radical prostatectomy. Eur Urol. 1999;35(5-6):399-407.
doi pubmed - Pathmanathan N, Balleine RL. Ki67 and proliferation in breast cancer. J Clin Pathol. 2013;66(6):512-516.
doi pubmed - Patil DT, Chou PM. Sialoblastoma: utility of Ki-67 and p53 as a prognostic tool and review of literature. Pediatr Dev Pathol. 2010;13(1):32-38.
doi pubmed - Sinn HP, Schneeweiss A, Keller M, Schlombs K, Laible M, Seitz J, Lakis S, et al. Comparison of immunohistochemistry with PCR for assessment of ER, PR, and Ki-67 and prediction of pathological complete response in breast cancer. BMC Cancer. 2017;17(1):124.
doi pubmed pmc - Zhang Q, Lou Y, Bai XL, Liang TB. Intratumoral heterogeneity of hepatocellular carcinoma: From single-cell to population-based studies. World J Gastroenterol. 2020;26(26):3720-3736.
doi pubmed pmc - Hammoud GM, Ibdah JA. Are we getting closer to understanding intratumor heterogeneity in hepatocellular carcinoma? Hepatobiliary Surg Nutr. 2016;5(2):188-190.
doi pubmed pmc - Kalasekar SM, VanSant-Webb CH, Evason KJ. Intratumor heterogeneity in hepatocellular carcinoma: challenges and opportunities. Cancers (Basel). 2021;13(21):5524.
doi pubmed pmc - McDonald KA, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, Opyrchal M, et al. Tumor heterogeneity correlates with less immune response and worse survival in breast cancer patients. Ann Surg Oncol. 2019;26(7):2191-2199.
doi pubmed pmc - An J, Oh JH, Oh B, Oh YJ, Ju JS, Kim W, Kang HJ, et al. Clinicogenomic characteristics and synthetic lethal implications of germline homologous recombination-deficient hepatocellular carcinoma. Hepatology. 2023;78(2):452-467.
doi pubmed - Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322).
doi pubmed pmc - Liu Z, Zhang Y, Shi C, Zhou X, Xu K, Jiao D, Sun Z, et al. A novel immune classification reveals distinct immune escape mechanism and genomic alterations: implications for immunotherapy in hepatocellular carcinoma. J Transl Med. 2021;19(1):5.
doi pubmed pmc - Wu SY, Liao P, Yan LY, Zhao QY, Xie ZY, Dong J, Sun HT. Correlation of MKI67 with prognosis, immune infiltration, and T cell exhaustion in hepatocellular carcinoma. BMC Gastroenterol. 2021;21(1):416.
doi pubmed pmc - Chida K, Oshi M, Roy AM, Yachi T, Nara M, Yamada K, Matsuura O, et al. E2F target score is associated with cell proliferation and survival of patients with hepatocellular carcinoma. Surgery. 2023;174(2):307-314.
doi pubmed pmc - Min Z, Xunlei Z, Haizhen C, Wenjing Z, Haiyan Y, Xiaoyun L, Jianyun Z, et al. The clinicopathologic and prognostic significance of c-Myc expression in hepatocellular carcinoma: a meta-analysis. Front Bioinform. 2021;1:706835.
doi pubmed pmc - Ferrin G, Guerrero M, Amado V, Rodriguez-Peralvarez M, De la Mata M. Activation of mTOR signaling pathway in hepatocellular carcinoma. Int J Mol Sci. 2020;21(4):1266.
doi pubmed pmc - Sun EJ, Wankell M, Palamuthusingam P, McFarlane C, Hebbard L. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines. 2021;9(11):1639.
doi pubmed pmc - Chen TI, Lee FJ, Hsu WL, Chen YC, Chen M. Association of dipeptidyl peptidase-4 inhibitors use with reduced risk of hepatocellular carcinoma in type 2 diabetes patients with chronic HBV infection. Cancers (Basel). 2023;15(4).
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


