| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Review
Volume 15, Number 2, April 2024, pages 149-168
Pigs: Large Animal Preclinical Cancer Models
Kirtan Joshia, b, c, Tejas Katamc, Akshata Hegdec, d, Jianlin Chengc, d, Randall S. Prathere, Kristin Whitworthe, Kevin Wellse, Jeffrey N. Bryanf, g, Timothy Hoffmana, Bhanu P. Telugue, Jussuf T. Kaifib, g, h, i, j, Satyanarayana Rachaganic, f, g, j
aDivision of Hematology & Medical Oncology, Department of Medicine, University of Missouri, Columbia, MO, USA
bSection for Thoracic Surgery, Department of Surgery, University of Missouri, Columbia, MO, USA
cRoy Blunt NextGen Precision Health Institute, University of Missouri, Columbia, MO, USA
dDepartment of Electrical Engineering and Computer Science, University of Missouri, Columbia, MO, USA
eNational Swine Resource and Research Center, Division of Animal Sciences, University of Missouri, Columbia, MO, USA
fDepartment of Veterinary Medicine and Surgery, University of Missouri, Columbia, MO, USA
gEllis Fischel Cancer Center, University of Missouri, Columbia, MO, USA
hInstitute for Data Science and Informatics, University of Missouri, Columbia, MO, USA
iSiteman Cancer Center, Washington University, St. Louis, MO, USA
jCorresponding Author: Jussuf Kaifi, Section for Thoracic Surgery, Department of Surgery, University of Missouri, Columbia, MO, USA; Satyanarayana Rachagani, Roy Blunt NextGen Precision Health Institute, University of Missouri, Columbia, MO, USA
Manuscript submitted November 7, 2023, accepted January 4, 2024, published online March 21, 2024
Short title: Pigs: Large Animal Preclinical Cancer Models
doi: https://doi.org/10.14740/wjon1763
- Abstract
- Introduction
- Porcine Models of Cancer
- Conclusion With Translational Scope of Pig Models in General
- References
| Abstract | ▴Top |
Pigs are playing an increasingly vital role as translational biomedical models for studying human pathophysiology. The annotation of the pig genome was a huge step forward in translatability of pigs as a biomedical model for various human diseases. Similarities between humans and pigs in terms of anatomy, physiology, genetics, and immunology have allowed pigs to become a comprehensive preclinical model for human diseases. With a diverse range, from craniofacial and ophthalmology to reproduction, wound healing, musculoskeletal, and cancer, pigs have provided a seminal understanding of human pathophysiology. This review focuses on the current research using pigs as preclinical models for cancer research and highlights the strengths and opportunities for studying various human cancers.
Keywords: Pigs; Swine; Oncopigs; Preclinical cancer model; Translational research; Genetic engineering
| Introduction | ▴Top |
Relevance and challenges of animal models in cancer research
Cancer remains the second leading cause of death worldwide [1]. In 2023, 1.9 million new cancer cases are expected to be diagnosed, of which more than 609,000 cancer-related deaths are projected to occur in the United States. By 2025, the incidence of cancer is expected to rise, with 19.3 million cases expected worldwide. Among cancers, lung and bronchial cancer remains the leading cause of cancer-related deaths and making up to 21% of total cancer cases, followed by breast (15%), prostate (11%), colorectal cancer (CRC, 9%), pancreatic (8%), liver (6%), and ovarian (5%) cancers [2]. Therefore, there is a great need for the development and investigation of new therapeutic and diagnostic modalities to combat cancer-related morbidity and mortality [3].
Using animals as models for cancer research has been a controversial topic. There is an emerging consensus that in vitro experimental studies do not necessarily recapitulate the tumor microenvironment. Therefore, xenograft mouse models, where human tumors are seeded into immune-compromised mouse models, are used as an alternative. However, these xenograft tumor models lack an intact immune system, which is crucial to reconstitute the tumor microenvironment. Consequently, genetically engineered mouse models (GEMMs) are used as an alternative for clinical research. However, most of the research results emanating from mouse models have not been translatable to humans due to variations in metabolism and gene expression profiles. In this regard, large animal models which have a close similarity to humans in terms of size, physiology and immunology are emerging as an alternative. Even though large animal models offer better replicability and are better models for human diseases, there still exists a disproportionate gap in the use of rodents over large animals for basic research [3]. This bias in the use of rodents as a foundational model for research is attributable in part to low costs of housing and husbandry, short generational interval, and the availability of tools and ease of generating custom genetic technologies, thereby elevating mouse as the prime model for biomedical investigation. However, the recent discovery and successful adoption of CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein) system in pigs and large animal models is facilitating genetic modification and generation of custom preclinical models with relative ease. Given the growing crisis in cancer load globally, multiple regulatory bodies overlooking the welfare of research animals and ethical concerns surrounding the use of non-human primates, and the poor translatability of rodent models, there is a need for revisiting the research paradigm and utilizing alternative models such as pigs for cancer research (Fig. 1). In this article, we will briefly highlight the advantages and considerations for use of pigs for cancer research and summarize representative examples of the use of pig models in cancer research.
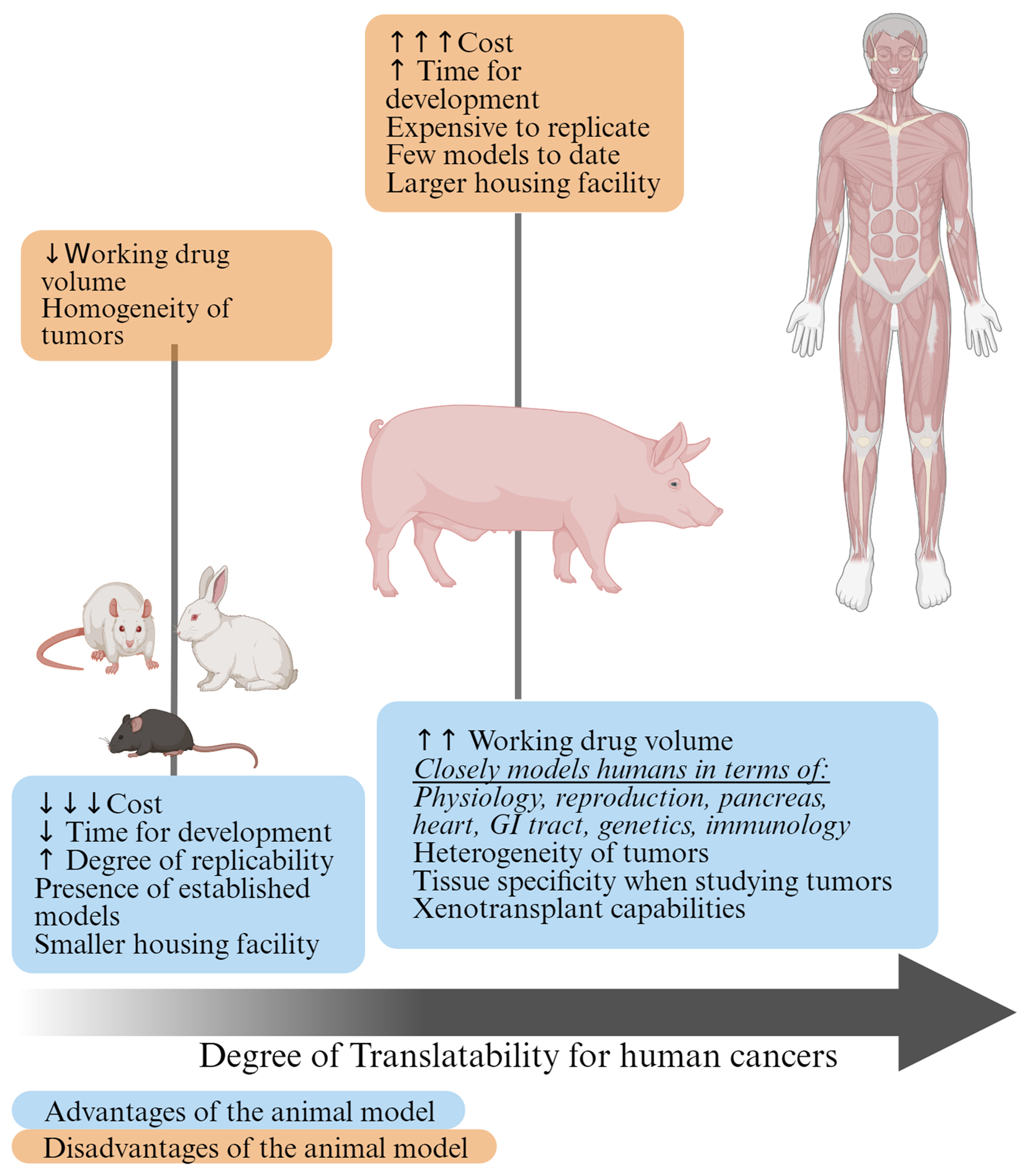 Click for large image | Figure 1. Advantages and disadvantages of using small and large preclinical cancer models. Given the similarities between pigs and humans, the pigs lie closer on the scale of translatability to humans compared to small animal models. |
Pigs as models for humans
The pig (Sus scrofa domesticus) was domesticated for agriculture by humans about 9,000 years ago in the Tigris Basin of Western Asia. However, the use of pigs as models for preclinical research is a recent phenomenon and still in its infancy. Approximately 730 pig breeds or lines have been generated worldwide, which range from miniature pigs to larger breeds that can reach up to 350 pounds. Pigs as biomedical models offer several advantages, which are highlighted below: 1) Piglets are precocial, can be delivered via cesarean section, and can be maintained in germ free isolators. Therefore, they are highly suitable for recapitulating human diseases. 2) Most pig breeds are outbred, and as such have greater heterogeneity, and best mimic outbred human populations [4]. As mentioned above, rodent models have historically been at the forefront of translational research by offering ease of access, handling, decreased generation interval, cost, imaging, and experimental capabilities. However, the most critical limitation of mouse models is the homogeneity with which mouse tumors originate compared to the heterogeneous and complex nature of human tumors [5]. 3) Mouse chromosomes are telocentric (having no obvious short arm at a cytogenetic level). A loss of heterozygosity event in a wild-type tumor suppressor gene allele can result in the loss of an entire chromosome in cells heterozygous for tumor suppressor gene mutation. Whereas in human tumors, loss of heterozygosity occurs via sub-chromosomal deletions covering the wild-type tumor suppressor gene locus. This recapitulates the differences between mouse and humans on a cellular level. 4) A greater similarity in coding regions of genes between pigs and humans along with chromosomal synteny, makes pigs better models to study the inception and progression of cancer [6]. 5) Additional advantages of pig models include: close similarity to humans in size, metabolism, gene expression profiles; greater than 80% similarity in immune parameters; availability of various breeds; relatively short generation interval; standardized and well-developed breeding conditions; larger litter size; advances in cloning and genetic engineering (GE) technologies; and are relatively cheaper and ethically more acceptable as models compared to primates [4]. 6) Additionally, pigs have been extensively used in surgical training such as robotic surgery, laparoscopic surgery, and other abdominal operations. Surgical trainees found working on live models improved their hands-on experience with human patients. Pigs having similar foregut anatomy to humans allows the surgical trainees to appreciate the tactile feedback of tissue and therefore reduces the chances of iatrogenic complications when working with human patients [7].
The annotation of the pig genome and the availability of large epigenomic and transcriptomic data sets has allowed precise analysis of tissue-specific regulatory activities. Independently conducted comparative analysis of pig, mouse, and human genomic data looked at categorizing orthologous genes based on the degree of conservation of gene expression between species (Fig. 2). Purely based on the width of the ribbons in the figure, mice might have more synteny (genes that lie on the same chromosome) with humans compared to pigs. Additionally, we conducted orthologous gene cluster comparison (via OrthoVenn3) between the genomes of humans, pigs, mice, rats, and dogs [8]. When compared to humans, pigs have 90 homologously shared elements, rats 17, mice 90, and dogs 103 (Fig. 3a, b). A pairwise heatmap was also generated to visualize the overlapping cluster numbers with the above-mentioned species (Fig. 3c). Similar analysis by another group yielded presence of more orthologous groups between humans and rodents compared to humans and pigs. Additionally, they conducted evolutionary breakpoint region analysis which yielded fewer breakpoints between human and rodents when compared to human and other mammals including dogs, pigs, and cattle. This would mean that rodents have diverged more recently than pigs from a common ancestor [9]. However, from our OrthoVenn3 [8] input investigating Expansion Contraction analysis, via CAFE5 method [10], revealed pigs to be closer to humans when comparing orthologous gene families. The expansion and contraction analysis is a method used in evolutionary biology and comparative genomics that can provide valuable insights into the evolutionary dynamics of gene families and the genetic basis of species-specific adaptations.
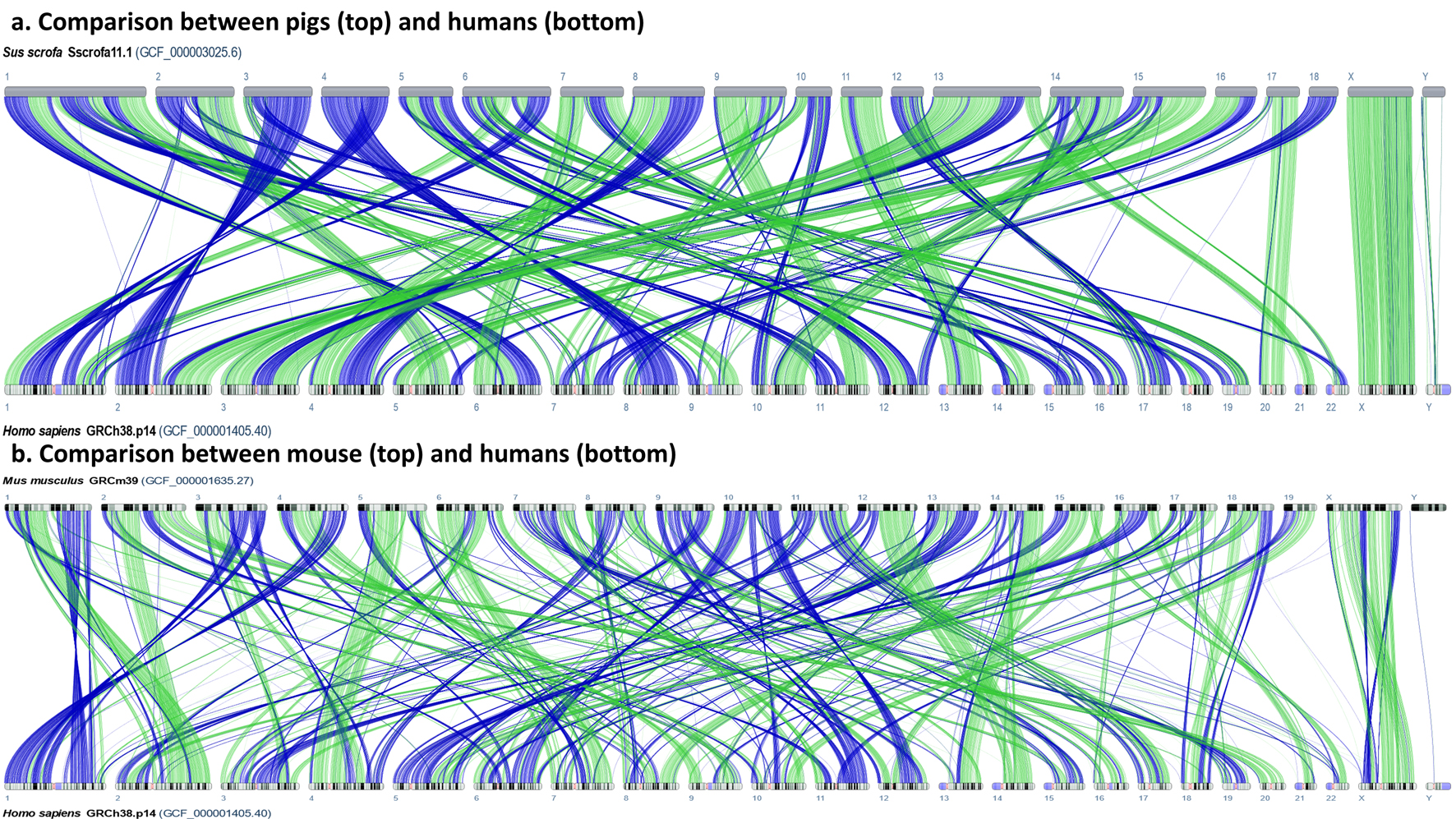 Click for large image | Figure 2. Comparative genome viewer alignment comparison between human-pig vs. human-mouse. (a) Comparison between pigs and humans. (b) Comparison between mouse and humans. The colored ribbons represent assembly-to-assembly alignment segments. These are reciprocal best placed alignments by default. Purple represents reverse alignments and green represents forward alignments only. Overall, the comparative genomic viewer shows synteny between the two species that are in comparison. Thicker bands are representative of continuous homology between those species. |
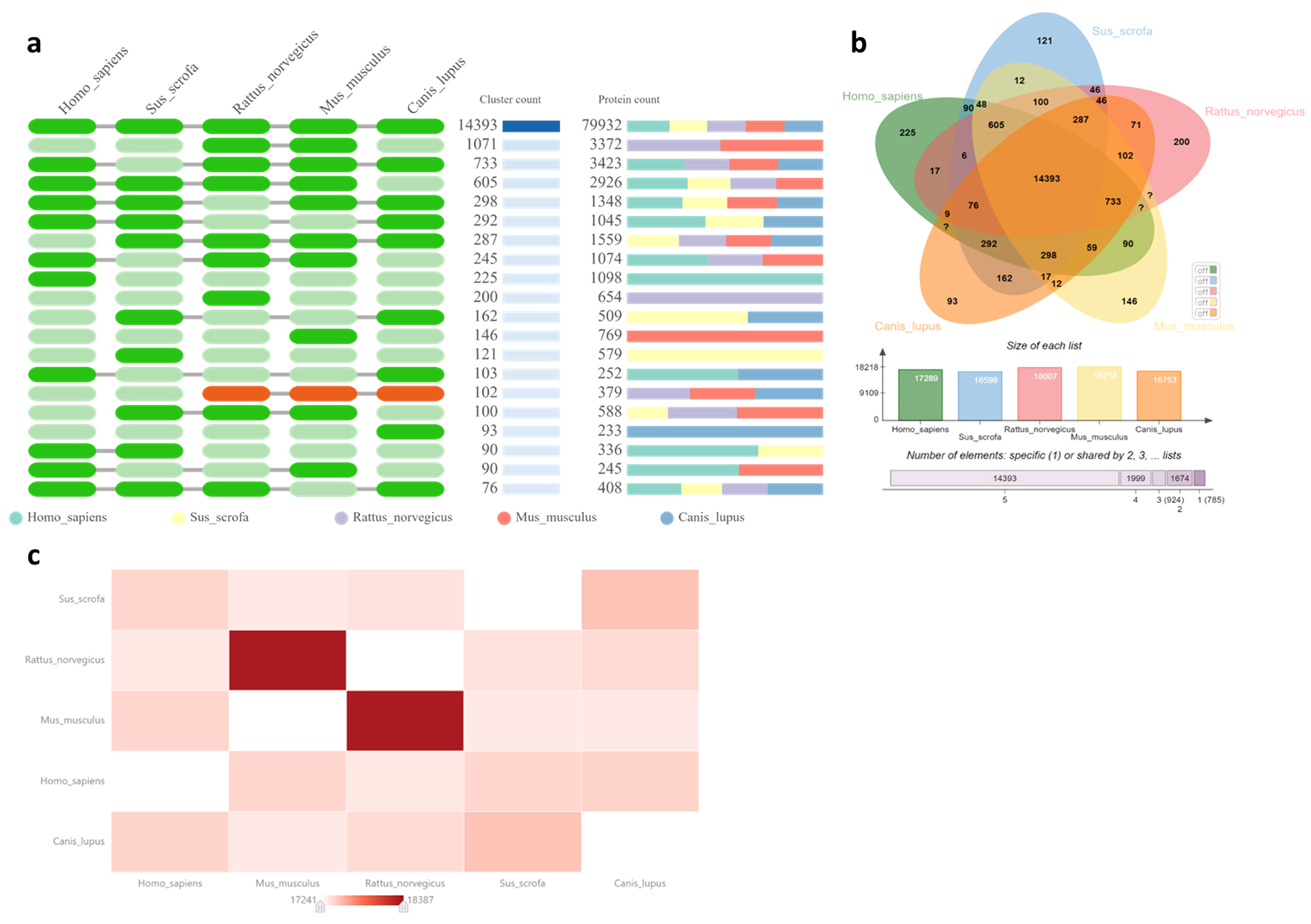 Click for large image | Figure 3. Orthologous analysis and comparison between humans, pigs, rats, mice and dogs. (a) Occurrence table presenting a unique number of shared homologous gene clusters among species along with the protein count on the right. (b) Venn diagram showing the number of shared orthologous gene clusters among species along with a bar graph comparison showing the number of orthologous clusters in each species. (c) Pairwise heat map comparison to visualize overlapping cluster numbers for five different species. Our orthologous analysis was conducted using default Orthofinder algorithm, with an e-value of 1 × 10-2, and inflation value of 1.50. |
This investigation included five distinct species: Homo sapiens, Sus scrofa, Mus musculus, Rattus norvegicus, and Canis lupus. Figure 4 shows the expansion and contraction analysis on the phylogenetic tree. It highlights that Homo sapiens and Sus scrofa, have diverged more recently, i.e., 1 million years ago, and are more likely to be epigenetically similar to each other when compared with Mus musculus that diverged earlier around 1.28 million years ago. While mice have more genetic synteny to humans, an emphasis should also be placed on epigenetic data when choosing an animal model to recapitulate tissue-specific human pathophysiology.
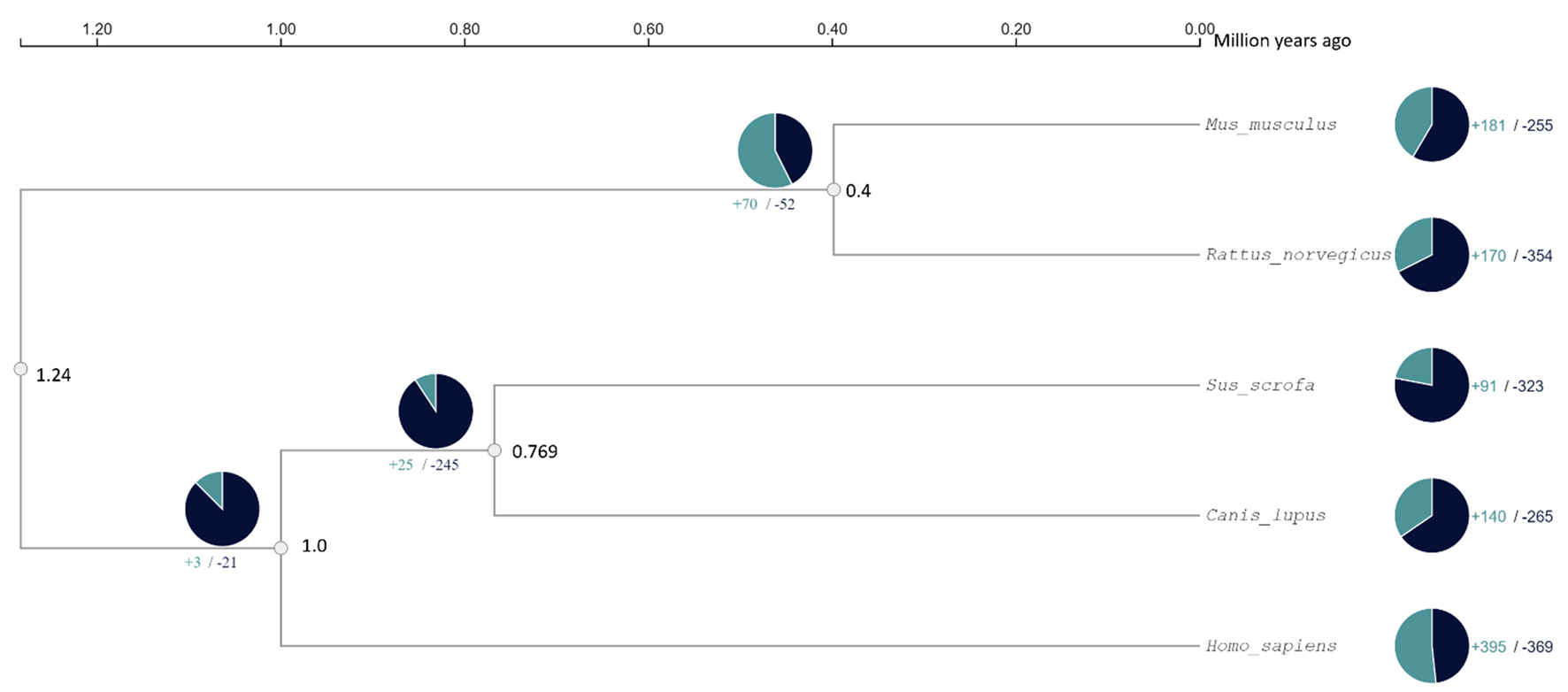 Click for large image | Figure 4. Expansion and contraction analysis of five species: mice, rats, pigs, dogs, and humans, respectively. In the pie charts, teal indicates expansions while black indicates contractions of orthologous gene clusters. |
Rodents also have elevated evolutionary rates compared to pigs. This could mean that pigs and human gene sequences would be more similar in comparison to humans and rodents. Rodent genomes are known to undergo large chromosomal rearrangements and accelerated mutational rates providing them with greater diversity than primates [11]. The ratio of non-synonymous to synonymous substitutions (dN/dS) is an important measure when evaluating the effects of natural selection on protein coding genes. Synonymous substitutions (nucleotide changes that do not alter the protein sequence) were estimated to be at least three times higher in rodents then other mammals which thus far has been explained by the shorter generation time of rodents compared to pigs or humans [12]. Despite the genetic data, epigenetic data gives a more detailed idea about choosing the right animal model when considering tissue-specific pathophysiology. Heritability enrichment analysis, measured by using stratified linkage disequilibrium score regression, estimates enrichment degree (e.g., the conserved enhancers divided by the proportion of single nucleotide polymorphisms (SNPs) of various complex traits mapped from pigs to similar regions in humans. This analysis of species-shared (preserved) chromatin states revealed higher enhancement for complex trait heritability when compared to the more species-specific (contrastive) chromatin states. Tissue-specific enhancers from human orthologous regions identified in pigs were also significantly enriched for the corresponding human complex traits which translated to similarity in biological functions of specific tissues in humans and pigs. Various enhancers in pigs were identified that showed significant enrichment of heritability of lung-, liver-, colon-specific, and brain cortex specific enhancers to corresponding complex human traits. This annotation of regulatory elements allowed precise comparison between human, mouse, and pig epigenomes in a tissue-specific manner enabling tailoring of specific models for specific diseases. For example, Alzheimer’s, Crohn’s and inflammatory bowel disease, adipose, body fat percentage, waist-to-hip ratio, and weight showed significantly enriched pig-to-human shared promoters and enhancers rather than mouse-to-human shared promoters and enhancers. Taken together, the comparative analysis of genomic and epigenomic data from mouse, pigs, and humans confirmed a high degree of conservation of genomic epigenomic elements, which only substantiates the argument that pigs may be a preferable alternative to rodents for translational research [13].
GE methods for generating transgenic pig models
In this article, GE refers to animals produced via genome editing and associated transgenic technologies. There are several review articles that highlight recent advances in genetic engineering and the readers are recommended to pursue them. Specific to the discussion below, Dmochewitz and Wolf reported an extensive timeline highlighting the evolution and advancement of GE techniques [14]. Lunney et al summarized recent GE pig models in use, along with a discussion of some of the most common editing techniques. In the past, most of the GE pig models were generated by pronuclear injection of zygotes or gene targeting in somatic cells followed by somatic cell nuclear transfer or cloning. Relatively recent advancements in CRISPR/Cas9 technology have fundamentally revised the research paradigm and has enabled simultaneous and multiplex genetic modification in pigs, which are enabling research in fields such as xenotransplantation. GE allows researchers, physicians, and agricultural scientists to genetically modify pig to study gene expression, disease progression, therapeutics, and production traits, respectively [15]. All GE pigs models discussed in the manuscript are tabulated in Table 1 [16-62].
 Click to view | Table 1. Summary of Pig Models |
| Porcine Models of Cancer | ▴Top |
Pig models for brain cancer
Traditionally, we have had a limited understanding of brain due to its high complexity, nuanced anatomy, ethical complications and technical difficulties in studying the organ [63]. In this regard, research using pigs has the potential to serve as a model due to similarities in cerebral structures and size of the brain between pigs and humans. The gyrencephalic pig brain allowed researchers to develop and perform advanced neuroimaging techniques leading to better translatability to humans [64]. Furthermore, transcriptomic, and protein-coding gene expression profiling, along with one-to-one gene orthologs comparison showed that humans, pigs, and rats exhibited evolutionarily preserved transcription factors which may provide understanding of brain architecture conserved through evolution. Human and pig brains exhibit high expression of glial fibrillary acidic protein (GFAP) and clusterin, which are biomarkers for neurodegenerative diseases such as Alzheimer’s disease and amyotrophic lateral sclerosis (ALS) compared to rats, while pig and mice brains have decreased levels of secretagogin (SGCN) - a protein expressed in the olfactory bulb, compared to humans [65, 66]. Once again placing importance on proteome profiles to choose an animal model most appropriate for translational research [67] to recapitulate tissue-specific needs.
Hicks et al [68] have performed meta-analysis on existing literature on pig glioma models. The first pig glioma study was conducted by Selek et al (2014) by injecting U87 GM and G6 human glioma cell lines into the parietal corona radiata via surgical implantation into pig brain. Subsequent magnetic resonance imaging (MRI) on these pig brains revealed 14 of 15 pigs had macroscopic tumors and displayed a mean volume change from the 15- to 28-day time interval after injection. Microscopically, the pigs had brain inflammation and mass effect on lateral ventricle and adjacent parenchyma. They also observed hypercellularity of the tumor showing infiltration of adjacent parenchyma, intense new angiogenesis and satellitosis with GFAP positivity showing astrocytic nature of the tumor. Successful tumorigenesis of human glioma cells in pigs showed their utility as a preclinical glioma model [16]. Khoshnevis et al (2017) studied glioma in Yucatan minipig models by injecting the minipigs with U87 GM cell lines and observed eight out of the nine pigs had developed significant tumors in a dose-dependent manner. These tumor cells were strongly positive for vimentin but negative for GFAP, which is contradictory to Selek et al [17]. A deeper dive into the understanding of patterns of gray and white matter functional network involvement in human glioblastoma multiforme (GBM) patients showed that there is some role in the involvement of the ventral frontoparietal tracts in predicting overall survival [69]. Therefore, the replicability of tumor induction in the parietal lobe of pig models enhances the importance of the data from indirect mapping of MRI scans of GBM human patients. In a recent study, Tora et al (2020) injected lentiviral vectors to induce high-grade glioma within minipigs instead of performing injections of human glioma cell lines. MRI scans on these pigs revealed mass lesions and subsequent immunohistochemical staining showed high levels of GFAP and OLIG2, indicators for high-grade glioma [18]. The anatomopathological study of glioma in pigs helped to develop novel drug testing delivery techniques, which would be influential in developing successful clinical trials in humans [19]. While studying brain pathology in humans is highly complex, pig models have attempted to bridge the gap in understanding pathophysiology of brain cancer and aided in developing novel drug delivery techniques. Therefore, large animal models show definitive promise to improve survival of human patients with high-grade glioma.
Osteosarcoma (OS)
Murine models are considered the predominant animal model for studying OS pathogenesis and treatment due to their small size and short generation interval. However, structural differences of rats and mice compared to humans create difficulties in studying OS and developing translatable therapeutics. Cancer biology of humans and rodents are also remarkably different for studying OS metastasis and genetic related events. Pigs and humans share similar lamellar bone structure, bone mineral density, and bone cross-sectional area. In addition, lengthy lifespans of pigs (12 - 15 years) also allow researchers to better understand OS progression [15, 70] and metastasis.
Sieran and colleagues (2014) developed a genetically engineered pig model by delivering a TP53R167H targeting vector to induce an R167H missense mutation in TP53 in fetal fibroblasts. Three pig lines were developed, TP53+/+, TP53R167H/+, and TP53R167H/R167H pigs. Using computed tomography (CT) and MRI, the group discovered no tumors developed in TP53+/+ and TP53R167H/+ pigs, while all sexually mature TP53R167H/R167H pigs developed neoplastic lesions (including lymphomas, osteogenic tumors, and a renal tumor) and two of seven developed osteogenic tumors. In one of the pigs developing an osteogenic tumor, the tumor migrated into the intracranial cavity and caused bone destruction, and in the other pig, an osteogenic tumor was identified in the long bone. The porcine TP53R167H mutation is orthologous to human TP53R175H mutation, making this porcine model a viable model to study human OS [20].
Saalfrank et al induced TP53 and KRAS mutations into porcine mesenchymal stem cells (MSCs) by introducing TP53R167H-/- -KRASG12D-/+ Cre-inducible alleles. MSC-excised CAG-loxp-stop-loxp (LSL)-TP53R167H homozygous-KRASG12D heterozygous and MSC-excised CAG-LSL-TP53R167H homozygous-KRASG12D heterozygous-MYC expression vector cells were xenografted into separate mice and MSC-excised CAG-LSL-TP53R167H homozygous-KRASG12D heterozygous injections resulted in one small nodule out of four injections while MSC-excised CAG-LSL-TP53R167H homozygous-KRASG12D heterozygous-MYC expression vector cells gave rise to three tumors out of four injections. These tumors were extracted and cultured to make porcine sarcoma cell lines, and these cell lines were injected into nine pigs. Out of the nine pigs, four pigs below the age of 16 months did not show any sign of tumors while of the five older pigs, four had tumors and one had ossifying lesions. Second filial generation CAG-TP53LSL-R167H/LSL-R167H transgenic pigs showed multifocal osteoblastic OS in multiple parts of the body. TP53-induced mutation caused heterozygous pigs to develop OS in an average of 20 months while homozygous TP53-mutated pigs developed tumors in as little as 7 - 8 months, thus showing the potency of TP53 mutation in OS. Like humans, the tumors mainly developed in the long bones [21].
Niu et al (2021) also used genetically engineered TP53-mutated pig models to understand OS function. Eighteen of the 29 heterozygous latent non-inducible (floxed, fl) TP53R167H pigs developed OS while all the homozygous flTP53167 pigs developed OS. The group identified high expression of both the Δ152p53α isoform, a highly upregulated isoform in organs vulnerable to cancer, and circTP53, responsible for increasing cellular proliferation of OS cells, in OS. The Δ152p53α isoform is encoded by P2 promoter, which is also present in humans, as the human P2 promoter encodes Δ160p53α. This indicates that the Δ152p53α isoform in pigs is equivalent to the Δ160p53α, which has been reported to be a highly expressed isoform in human cancers [22]. Being able to modulate specific epigenetic elements further underscores the need for understanding epigenomic and transcriptomic data sets before committing to a large animal model for future translational research.
Hematological malignancies
Hematological malignancies, which include leukemia, lymphoma, and myeloma are projected to account for 184,270 deaths in the United States in 2023, and while mortality rates have declined, creating better therapeutic options is of utmost importance [2]. Although the murine model is relatively inexpensive compared to the pig model, the numbers of genetic, immunologic, and physiologic differences between mice and humans are vast. Pigs and humans share similar immune and lymphocytic profiling. Pigs, however, possess greater levels of gamma delta T cells than humans. Pigs and humans share similarities of lymphohematopoietic malignancies such as chronic myelogenous leukemia (CML), severe combined immunodeficiency (SCID), host-derived T-cell lymphoma and chronic lymphocytic leukemia (CLL). The development of CML in pigs and humans is associated with the defects of the nucleoporin gene, as NUP107 leads to CML in pigs while NUP98 leads to CML development in humans [71]. SCID pigs have non-existent T- and B-cell levels and two mutations in the Artemis gene, like human SCID patients, recapitulating the similarities in pigs and humans. Host-derived T-cell lymphoma and CLL arose in two SCID pigs after bone marrow transplantation, and the development of both malignancies may be linked to a “leaky” Artemis gene, which has been well characterized in human SCID patients. Therapeutics for hematological malignancies have been effective but pose fatal side effects. Pig models have tried to close the gap in this regard. Pig models have been used to test cellular therapies such as, donor leukocyte infusions in immune tolerant swine chimeras [23], and natural killer (NK) cell therapies in vitro in SCID pigs [24], in grafts vs. host disease (GvHD) and acute myeloid leukemia (AML) [72].
Duren-Struuck et al (2010) studied spontaneous CML in five inbred Massachusetts General Hospital major histocompatibility locus (MGH-MHC) pigs by performing blood counts via peripheral blood smears of the pigs. They observed acute levels of leukocytosis, mild anemia, and upregulated levels of lactate dehydrogenase (LDH). Gross pathology showed hepatosplenomegaly in all pigs. Splenic hemorrhage and widespread nodules in the liver were observed in all pigs, and in vitro culture of the tumor showed abnormally pale bone marrow via sternal biopsy, thus highlighting a leukemic process. Histopathological assessment showed elevated myeloid-to- erythroid ratios, low levels of metarubricytes, increased levels of eosinophils and myeloid precursors in bone marrow, and infiltration of malignant cells in lymph nodes and kidneys, all of which are human CML histopathological characteristics. Cell lines developed from these pigs are similar to human tumor cell lines in which there is a Philadelphia (Ph+) chromosome mutation and BCR-Abl fusion, a mutation highly represented in human CML. This evidence further underscores genetic similarities between human CML and swine CML [25].
Matar et al (2015) injected MGH-MHC miniature pigs with hematopoietic cell transplantations (HCTs) to test for the development of post-transplantation lymphoproliferative disorder (PTLD) and its translatability to humans. They observed 34.4% PTLD incidence in the pigs and the group described the effects of mobilization and T-cell depletion, cyclosporin A (CyA) treatment, and lymphocyte depletion on pigs with PTLD incidence. The PTLD incidence rate of pigs receiving granulocyte colony-stimulating factor (G-CSF) growth factors that make bone marrow and produce white blood cells, versus pigs receiving stem cell factor/interleukin 3 (SCF/IL3) mobilized cells, important regulators in hemopoietic cell development, was of no significance. Similarly, pigs that received T-cell depleted cell lines, as compared to those that did not, also saw no increase of PTLD incidence. The length of CyA treatment, lymphocyte deletion, and MHC mismatch also had no effect on the development of PTLD. Furthermore, the group also observed elevated levels of LDH in pigs having PTLD compared to those that did not, which is similar to humans. Previously the group reported increased incidence of PTLD after HCT in swine that have been conditioned with thymic irradiation, CyA, and T-cell depletion after transplantation. A recent study revealed that replacement of thymic irradiation with total body irradiation, resulted in similar numbers of B cells early post-transplantation, greater number of T cells at day 0 and an overall decrease in PTLD incidence. They concluded that a threshold number of T cells (and potentially memory T cells) are necessary to prevent further B-cell proliferation and therefore the development of PTLD in swine [26]. LDH has been used in humans as a marker for myeloid leukemias. This model recapitulated LDH can also serve as a supportive diagnostic marker for swine PTLD lymphomas. The increase in LDH could be detected at least 1 - 2 days in advance before an increase in WBC count and well before any clinical manifestation of lymphadenopathy [26]. Therefore, this model can help to study the incidence of PTLD within a short period of time, which is highly advantageous for translational research capabilities.
Boettcher et al (2020) used a CRISPR/Cas9 site directed ART-/- IL2RG-/Y SCID pigs that lack T cells, B cells, and NK cells and created ART-/- IL2RG-/Y fetal fibroblasts. Somatic nuclear transfer was performed by using these fibroblasts and the embryos were transferred into surrogate gilts, and a C-section was performed to deliver the ART-/- IL2RG-/Y mutated piglets. As expected, these piglets lacked T cells, B cells, natural killer (NK) cells, and lymphoid organs. Bone marrow transplant on an ART-/- IL2RG-/Y male pig restored T and NK cell levels in pig’s blood. To test whether ART-/- IL2RG-/Y pigs could engraft human CD34+ hematopoietic stem cells, they injected CD34+ stem cells into a pig fetus and found human cells circulating in peripheral blood and restored the bone marrow [73]. Therefore, the compatibility of CD34+ between humans and pigs offers a potential therapeutic method. In addition, CD45+ cells, which are highly expressed in hematological malignancies [74], as they regulate T cell levels, were found in ART-/- IL2RG-/Y pig bone marrow, liver, spleen, and thymic tissue [27]. Given immunological similarities between pigs and humans, pigs can serve as a viable and robust model to study human hematological cancer.
CRC and gastrointestinal (GI) conditions
There are many prominent similarities between the human and pig GI tract [75]. For example, the average nutritional requirements and mineral absorption, and gallbladder function are highly similar; and approximately 96% similarity has been found in microbiome functional pathways. The general physiology, hemodynamic parameters, vascular remodeling mechanisms, common absorption, distribution, metabolic, and excretion mechanisms are also similar. The complex interplay between immune system and GI tract also holds many similarities [15] between humans and pigs. However, some differences exist between pig and human GI tract such as the presence of a spiral colon, difference in pressure-diameters of bile ducts [76], inverted lymph node structure, distribution and frequency of intestinal lymphocyte populations, and the presence of a continuous ileal Peyer’s patch [75].
Pigs and humans both use the colon for water and electrolyte absorption. The colonic muscle layer helps in removing waste from the body, and microbiome in the colon further breaks down undigested waste material to produce bacterial metabolites. These metabolites are important for human and pig physiological processes. Both species also have similar digestion time and processing of micro- and macronutrients. However, porcine GI microbiota is different than humans in terms of butyrate producers. With the advancement of gnotobiotic models there has been successful establishment of microbiome inocula in pigs from infant human gut using Bifidobacterium and Bacteroides. Accordingly, pig models are increasingly being used to study the pathogenesis of various human illnesses like gastritis, gastric ulcers, and predisposition to cancer secondary to Helicobacter pylori, the effects of formula supplementation versus breastmilk feeding, necrotizing enterocolitis, short-bowel syndrome, metabolic syndrome, and changes in diet resulting in obesity, inflammation, and metabolic diseases [15].
Pigs bearing a 93-bp deletion of the adenosine-uracil-rich element (ARE) and a constitutive-decay element within the 3′ untranslated region of the TNF gene (TNFΔARE) are being used to mimic the pathophysiology in inflammatory bowel disease patients. The group established pigs that had varying levels of inflammation intensity within the GI tract. Immunohistochemistry showed increased Ki-67 and leukocyte and lymphocyte infiltrations (IBA1, CD3), and decreased number of mucus-secreting goblet cells (PAS/AB+). Increased intestinal inflammation also yielded bacterial dysbiosis in the mutated pigs. TNFΔARE pigs show promise in recapitulating the major characteristics of human inflammatory bowel disease [28]. A high-calorie diet is a potential risk factor for colon cancer and type 2 diabetes. A human-relevant pig model was used to show that a high-calorie diet leads to increased expansion of proliferative and stem cell zones in the pig colon. In particular, two distinct stem cell populations (ASCL-2 and BMI-1) were observed in pig colon, as is similar to humans, upon exposure to a high-calorie diet. However, these findings were not observed in mice [77]. This further substantiates the claim that pigs are a superior model in terms of translatability to humans.
Mutation in adenomatous polyposis coli (APC) is orthologous to the germline mutations present in patients with familial adenomatous polyposis (FAP) condition. When comparing pigs to humans, there is a similar distribution of early intestinal polyposis due to APC1311, starting from the cecum to the rectum. The difference is evident when comparing mice and humans. Murine (Apcmin) is mostly localized to the small intestine with some present in the distal colon. Additionally, there are similarities between the development of colonic, rectal polyps and adenomas in genetically modified pigs and in human FAP and CRC patients. For example, in the APC1311 pig model, expression of the truncated APC along with reduced expression of wild-type allele enhances cellular proliferation and increases the risk of secondary mutations, such as a loss of heterozygosity resulting in polyposis. This supports the data that reduced APC mRNA expression is associated with polyp formation and human FAP patients. Therefore, it may be deduced that APC can epigenetically act as a FAP modifier gene. APC expression imbalance then may be labeled as a disease risk factor [29, 78].
The similarity in anatomical size between the pig and human GI tract also allows for testing and advancing endoscopic techniques, particularly for colonoscopies. In fact, the pig APC1311 model has been used in the development of cathepsin protease-activatable probe for fluorescence-guided endoscopy [79], fluorescent silica nanoparticles-guided detection of colorectal adenomas using video-rate fluorescence-assisted white-light endoscopy [80], and to train artificial intelligence to detect more adenomas [81]. Colonoscopies are imperative in the early detection of colonic dysplasia [30, 75]. Furthermore, a recent study has indicated the utility of pig as a translational intestinal epithelial stem cell (ISC) research model. CRISPR/Cas9 editing was used to develop a transgenic porcine leucine rich repeat containing G protein-coupled receptor 5 (LGR-5), LGR-5-H2B-GFP pig model. Intestinal epithelial stem cells from this model were also used to develop a CRC organoid platform. The importance of the porcine model was underscored by its expression of olfactomedin-4 (OLFM4), and three additional stem cell markers that are similar to humans ISCs in the small intestine and colon. In contrast, in mouse Olfm4 expression is restricted only to the small intestinal ISCs. Therefore, an OLFM4 producing porcine model could provide important data for modeling CRC where OLFM4 is a known marker for cancer cells and potential metastasis [30]. Schaaf et al (2022) extensively reviewed available models of CRC such as TP53R167H and KRASG12D [75].
Pancreatic cancer
Pancreatic cancer continues to be one of the deadliest cancers with grim prognosis, with incidence to mortality ratio of nearing 1. It is the third leading cause of cancer-related deaths in both men and women and projected to be a second leading cause of cancer-related death by 2030 [2]. It has one of the lowest 5-year relative survival rates (10%) compared to other cancers. Depending upon the stage of diagnosis, distant metastatic pancreatic cancer patients have a 5-year survival rate of 3% [82]. Pancreatic cancer and its metastasis interestingly behave oddly. Tagging and tracking pancreatic epithelial cells in a mouse model revealed that tagged cells invaded and entered the bloodstream unexpectedly early, before a malignancy could be detected by histological analysis. These pancreatic circulating cells interestingly maintained the mesenchymal phenotype, had stem cell properties, and were found to have seeded the liver. Systemic inflammatory events leading towards foci formation or pancreatitis resulted in an increased number of circulating pancreatic cells. Interestingly dexamethasone at this time was shown to be immunosuppressive and prevented mesenchymal pancreatic cell seeding [83]. The KPC (Pdx-Cre x CAG-LSL-KrasG12D-Trp53R172H) mouse has been the gold standard for pancreatic research for over a decade [84]. However, there are large anatomical and physiological variances between human and mouse pancreas. While the human pancreas is a retroperitoneal and segmented organ, the mouse pancreas is diffuse, dendritic, and poorly lobulated. Furthermore, the secretion pathway of the exocrine pancreas and the endocrine component are significantly different between mice and humans. Mice have a relatively larger percentage of insulin producing beta cells, whereas humans have a larger proportion of glucagon-producing alpha cells [3]. In contrast, human and pig pancreases have similar development and morphology patterns, 99% homology in insulin amino acids sequence, and islet cells being dispersed throughout the exocrine pancreas [15]. Due to the significant differences between the mouse and human pancreas and the above-mentioned similarities between the pig and human pancreas, pigs are considered as a more suitable model to studying pancreatic cancer and is superior in comparing drug performance/toxicity for translation to humans.
Principe et al [31] created the first large animal model for pancreatic carcinogenesis. In this study, 7 days post-transduction with adenovirus Cre (AdCre) into isolated pig pancreatic duct cells, they were able to identify duct cells that had spindle-shaped morphology consistent with malignant transformation. The transduced cells strongly expressed E-cadherin (indicating epithelial origin) and CK19, affirming their ductal lineage, displayed increased RAS activation, KRAS effector pERK1/2 and PCNA (proliferation surrogate), thus confirming the tumorigenic phenotype. Implantation of transduced cells subcutaneously into SCID mice resulted in several large masses. Additionally large focal plaques on the abdominal viscera with intraperitoneal injection were also observed. These tumors in mice had overall similarity to human pancreatic ductal adenocarcinoma (PDAC) and were able to show metastatic behavior. Restriction of AdCre delivery to the main pancreatic duct in vivo led predominantly to a PDAC exocrine histotype. Interestingly, 1 year following AdCre injection, they had no overt signs of illness or pancreatic insufficiency, no evidence of gross tumor formation on contrast-enhanced computed tomography (CT), and unremarkable regional lymph nodes. However, upon further dissection of the main pancreatic duct, they developed several large nodular tumors with a pronounced fibrous component at the site of AdCre cannulation. These lesions were consistent with the previously mentioned PDAC phenotype. They also found several separate areas with phenotypes similar to pancreatic neuroendocrine tumors (PNETs), while displaying less pronounced desmoplastic tumor stroma and positivity for neuroendocrine marker, synaptophysin. Both the exocrine derived tumor and the neuroendocrine type of tumor displayed signs of excessive proliferation. The porcine PDAC, showed dense desmoplastic tumor stroma, tumor associated fibrosis and mesenchymal origin with vimentin positivity. Considering the similarities in pancreatic carcinoma between pigs and humans, pigs turn out to be a better model for studying PDAC than are mice.
A cell line from the pancreatic duct of domestic pigs was transformed by using oncogenic KRAS and Simian virus 40 (SV40T) and grows tumors when injected subcutaneously into nude mice [85]. Additionally, an inducible negative regulator protein (rtTR-KRAB) pig model has also been generated by using primary porcine fibroblasts with murine Pdx-1 promoter overexpression of oncogene cassette containing MYC, KRASG12D and SV40 LT. This model specifically allows on-and-off expression via doxycycline which could repress the expression of the three oncogenes. By using this model, increased phosphorylation of ERK was confirmed due to the constitutively activated mutated K-rasG12D in murine Pdx-1 expressing cells during pancreatic organogenesis. Similar to the study by Principe et al, the size and morphology of the pancreas appeared normal when compared to the wild-type pigs. Hyperplastic foci of acinar cells were found to be localized with increased cell density in the neonatal pancreas 45 days post farrowing. However, progression of these hyperplastic foci to cancer and metastasis needs to be better characterized in mice and humans. However, this model can provide valuable insights into the initial stages of pancreatic carcinogenesis. In addition, this models’ ability to repress the expression of commonly observed oncogenes could help in studying pancreatic cancer progression [32] and identify the oncogenes as a molecular target.
Mondal et al [33] utilized two different techniques of AdCre induction of pancreatic cancer in transgenic Oncopigs (CAG-LSL-KRASG12D-IRES-TP53R167H). In the first technique two Oncopigs were injected with AdCre into the main pancreatic duct plus an injection into the parenchyma of the duodenal lobe; however, there is no gross microscopic evidence of tumors observed in these Oncopigs. By using the second technique, AdCre was injected into the duct of the connecting lobe in 12 Oncopigs. Out of which only two Oncopigs lived until the planned euthanasia date, whereas the other 10 remaining Oncopigs underwent unplanned euthanasia due to lethargy, respiratory distress, and progressive decline of health. These 10 Oncopigs had peripancreatic phlegmon. Upon further inspection the phlegmon yielded abundant tumor cells with large nuclei and prominent nucleoli. Immunohistochemistry from the peripancreatic phlegmon was positive for mutant KRASG12D (70% quantification) and mutant p53 (40% quantification). Stained tumor sections were significantly positive for Ki-67, Alcian blue staining, vimentin, and CD31. Additionally, hallmark EMT genes were up-regulated in the tumor sections of Oncopig compared to wild-type. Analysis of the tumor microenvironment also showed that transforming growth factor beta (TGF-β) signaling, matrix metalloproteinases (MMP), interleukin 18 (IL-18) signaling and TH17 cell differentiation signaling pathways were significantly up-regulated in Oncopigs compared to wild-type. Additionally, similar to human PDAC, Oncopigs pancreatic tumors also had overexpressed MMP1, MMP3, MMP12, MMP19, TIMP1, ITGB3, FN1, interleukin 27 receptor subunit alpha (IL27RA) and transforming growth factor beta receptor 1 (TGFBR1) genes. Together these findings suggested that porcine pancreatic tumors had similar characteristics to the human PDAC. Pancreatic tumors formed with an incidence of 71%. These tumors were predominantly epithelial in histology, and less differentiated based on gene expression. Interestingly, due to the similarities between humans and pigs in terms of translatability, porcine urinary bladder (PUB) even has been used as an advanced organ culture model for shaping an ex vivo pancreatic niche. In this case, the ex vivo model offers an earliest platform for pancreatic dysplasia and cancer if the implanted pancreatic duct like organoids feature KRASG12D mutations [86]. This further substantiates the claim of pigs as a superior model for studying pancreatic cancer as compared to other model organisms.
Hepatocellular carcinoma (HCC)
Liver cancer is the fifth leading cause of death in men and seventh leading cause of death in women. Liver cancer-related mortality rates have significantly increased for decades; however, rates have stabilized in women and have begun to decline in men from 2017 to 2020 [2]. Only a minority of the early-stage liver cancer patients are eligible for surgical resection. However, most patients are diagnosed in advanced stages, and the treatment options become limited. Considering treatment, transarterial chemoembolization resulted in 23% improvement in 2-year survival. Kinase inhibitors, such as sorafenib, became the most widely accepted option for late stages. Transplantation can be potentially curative; however, not all patients are surgical candidates. This underscores the need for improved understanding for HCC pathogenesis, detection, and treatment strategies [87]. The Oncopig HCC model can bridge this gap due to similarities in epigenomic and transcriptomic data between pigs and humans [13].
Schachtschneider et al [34] have developed and characterized the Oncopig as a model for human HCC due to similarities in phenotype, gene expression, and tumor development. The Oncopig primary hepatocyte cell lines were exposed to AdCre in vitro leading to activation of mutant KRASG12D and TP53R167H transgenes. The resulting HCC cell line displayed similar set of pathological characteristics to human HCC cells including nuclear hyperchromatism, pleomorphism, increased nuclear to cytoplasmic ratio, and round to oval pale eosinophilic or granular cytoplasm and 80% of the Oncopig HCC cell line expressed vimentin indicating epithelial-mesenchymal transition. The HCC cells are also able to secrete alpha-fetoprotein (AFP), a serum marker that has been commonly observed in majority of human HCC. Injection of Oncopig HCC cells subcutaneously into the hepatic parenchyma of SCID [35] mice resulted in formation of tumors within 21 days. These tumors were histomorphologically similar to human HCC, exhibited linear growth curve, and displayed significant angiogenesis. Autologous subcutaneous transplantation of Oncopig derived HCC cells showed a palpable mass dependent on the dose of Oncopig HCC cells injected. These tumors are Edmondson-Steiner grade 2 human HCC with a trabecular patterning. Oncopig HCC cells gene expression profiling revealed activation of TP53 dependent cell cycle progression, expression of pro-angiogenic factors, evasion of apoptosis, activation of the telomeric maintenance, and subclass specific Wnt signaling activation. Additionally, they observed a high level of variability in the regulation of gene expression (elevated compared to reduced) in human HCC cell lines. Although master regulators (transcription factors that play an important role in gene expression) driving increased gene expression were observed in Oncopig HCC cell line. The same master regulators were not identified in human HCC cell lines. However, they were able to identify eight transcription factors that had reduced expression in both Oncopig and human HCC cell lines. To further add to the importance of pig as a preclinical cancer model, tumor infiltrating lymphocytes were identified in the Oncopig HCC tumors indicating that these are “hot” tumors underscoring its relevance for immunotherapy trials.
Additionally, they were able to replicate cirrhosis development in Oncopigs via a transarterial alcohol injection [34, 36]. They were able to appreciate reproducible liver gene expression profile changes post transarterial ethanol exposure. Since alcohol consumption is a major cause of liver cirrhosis and it also increases the risk for HCC development, therefore, developing Oncopig models would be beneficial to study alcohol-mediated HCC pathogenesis. Similar to human patients’ chronic alcohol exposure is necessary for fibrogenesis. However, down-regulation of MYC in Oncopig fibrotic liver samples was observed compared to the elevated expression observed in human cirrhotic livers. MYC overexpression leads to hepatocyte proliferation, activation of hepatic stellate cells, and induction of fibrogenesis. Reduced MYC expression in this case suggested that hepatic stellate cells are no longer being activated 8 weeks post induction of ethanol as there was no chronic alcohol exposure, meaning there was a subsequent down-regulation of MYC expression and resolution of fibrosis indicating liver recovery. They also showed chemotherapeutic-based hepatotoxicity and reduced chemotherapeutic tolerance replicated in Oncopigs similar to humans, secondary to impact of alcohol exposure on the expression of drug metabolizing enzyme gene families, yet again presenting evidence that pigs are a robust model to replicate human conditions [36].
Gaba et al [35] went on to describe similarities in treatment response between pigs and humans. Similar expression of uptake transporter SLC22A1, the efflux pump ABCB1, and UGT1A1, drug metabolizing enzyme were seen between Oncopig and human HCC cell lines. Phase 1 sorafenib metabolizing enzyme CYP3A4 (porcine homologue CYP3A39) was reduced whereas CRB1, involved in doxorubicin metabolism, had increased expression in Oncopigs compared to human HCC cell lines. When testing for cell lines chemotherapeutic susceptibility to locoregional and systemic HCC treatment, Oncopig HCC cell line response was more predictive of human HCC response compared to murine HCC response. This underscores that Oncopig model can potentially serve as a bridge between murine and human studies. Concurrent liver fibrosis induction prior to the engraftment of subcutaneous tumor fragments into the liver of Oncopigs led to the induction of reproducible Oncopig HCC tumors in vivo. Additionally, whole genome sequencing of Oncopig intrahepatic HCC tumor showed intratumor heterogeneity and single nucleotide variants indicating accumulation of somatic mutations in distinct tumor cells as commonly observed in human HCC. The CRISPR/Cas9-mediated knockout of Oncopig TP53R167H showed reduced cell proliferation compared to the parental line. Given the resemblance between Oncopig and human HCC at the genomic level, while showing that Oncopig HCC cell lines can be genetically manipulated to tailor HCC tumors, pigs are a meaningful model for investigating clinically relevant cancer phenotypes and testing new precision medicine modalities.
Elkhadragy et al [37] studied the applicability of CRISPR/Cas9 and effect of different mutational profiles on tumor progression and chemotherapeutic susceptibility. The AT-rich interactive domain-containing protein 1A (ARID1A), subunit of SWItch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex, which is mutated in several types of cancer, including HCC, is shown as a tumor suppressor gene. In this study, authors introduced loss of function mutations in porcine ARID1A through Cas9-mediated nonhomologous end joining method. Human and porcine ARID1A gene sequences showed 89% identity and coding regions revealed 95% identity. Same group [38] developed a simultaneous knockout of (loss of function mutation) ARID1A and AXIN1, another commonly mutated gene HCC. There was no appreciable effect of this dual knockout on the susceptibility of the most used human HCC treatments, sorafenib and doxorubicin, in porcine HCC cells. Orthologous injection of the dual edited HCC cells resulted in the development of subcutaneous tumors in Oncopigs.
Pigs have also been used in therapeutic trials. In one such study, hereditary tyrosinemia type-1 (HT1) [39] and its treatment with 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) is compared against in vivo administration of lentiviral vector targeting the expression of human fumarylacetoacetate hydrolase (FAH). As a result of FAH deficiency in patients with HT1, there is an accumulation of toxic metabolites in the liver resulting in oxidative damage. Patients usually develop fibrosis, cirrhosis, high rates of HCC, and early onset of liver failure, if untreated. Current pharmacological treatment with NTBC reduces the formation of toxic metabolites by inhibiting upstream enzyme 4-hydroxyphenylpyruvate dioxygenase (HPD) converting HT1 to its benign phenotype. Despite the available treatment of NTBC to patients, they still develop long-term complications and to date the only curative treatment is liver transplantation. Mice models have failed to recapitulate this disease due to their propensity to independently develop fibrosis and cirrhosis, unlike pigs and humans. A minimally invasive approach using percutaneous ultrasound-guided portal vein administration of lentiviral vector caring human FAH was trialed in FAH-/- pigs. NTBC-independent weight gain was observed with correction of liver markers after lentiviral FAH delivery along with newly repopulated FAH-positive hepatocytes. AFP also showed an age-dependent decrease over time in the animals treated with lentiviral FAH compared to untreated pigs. Portal vein delivery was superior compared to systemic delivery of lentiviral FAH. Portal vein delivery resulted in positive vector presence in all lobes of the liver by day 337, whereas systemic delivery (via ear vein injection) resulted in no detection of vector presence in the liver but was observed in spleen, pancreas, duodenum, and lung at 14 days post-infusion. Next generation sequencing and bioinformatic analysis of hepatocytes at the time of euthanasia revealed a benign integration profile after in vivo delivery of lentiviral FAH. No enrichment of tumor-related pathways was found upon gene set enrichment analysis. While the animals with intervention were cycled on- and off- of NTBC, learning from pig model, an argument can be made for chronic low-dose therapy of NTBC in humans. Most impressive feature is the absence of chronic inflammatory changes in the liver post lentiviral therapy. This is protective towards the formation of fibrosis, cirrhosis and eventually HCC. In fact, restoration of wild-type phenotype was observed by 12 months post treatment, with undetectable AFP levels at 1 year. While the study used a small number of animals, the results at least make the argument that pigs can be used for translational clinical research that can directly impact humans.
Bladder cancer
Bladder cancer (urothelial cancer of the bladder) is one of the most common malignancies affecting the urinary system. Bladder cancer has been divided into two different categories: non-muscle and muscle invasive. Current treatment for non-muscle invasive bladder cancer includes transurethral resection of bladder tumor, intravesical mitomycin C or with bacillus Calmette-Guerin (BCG). Treatment for muscle invasive bladder cancer includes radical cystectomy, systemic chemotherapy, or local radiation therapy. Bladder cancer is highly resistant and prone to relapse despite the availability of current therapies. There is an unmet need to better understand the pathophysiology of bladder cancer, identification of diagnostic and prognostic markers and to develop new therapies. For these goals a reliable and clinically translatability animal model for bladder cancer is imperative [88, 89]. A summary of mouse models for bladder cancer is extensively reviewed in previous studies [40, 88]. Due to the overall loss of heterogeneity in mice bladder cancers compared to human bladder cancers, rare incidence of spontaneous bladder cancers in mice, replicability issues, and length of time needed for tumor induction, mice are considered inferior for translatability to human bladder cancer research [89]. A comparative large animal model can potentially fulfill the unmet need to better understand the pathophysiology of bladder cancer. A cryocatheter performance for the treatment of bladder cancer was also investigated using pigs [90].
In patients with radical cystectomy, ileal neobladder construction is a common treatment. GI segments remain the primary source for tissues used for urinary tract reconstruction. As a result of the transposed intestinal segments, patients usually have metabolic acidosis early postoperatively. Additionally, patients have increased mucus production by the transposed intestinal segments, resulting in recurrent urinary tract infections, gradual renal dysfunction, urinary retention, and even neobladder perforations. Recently a group tried bladder tissue engineering as a promising alternative approach for neobladder construction in a porcine model. Autologous peritoneal graft consisting of a peritoneal sheet and the seromuscular layer of the ileum were used as the reconstructed neobladder. This showed normal function and overall better gross morphological characteristics. Urothelium-like cells expressing urothelial biomarkers also appeared in the neobladder. Peritoneal tissue was chosen due to its abundance and proximity, and seromuscular layer of the ileum due to its easy adaptability, sufficient elasticity, and mechanical strength. Interestingly, the control ileal neobladder retained its gross morphological appearance to the ileum with abundant folds, whereas the luminal surface of the neobladder was smooth. Peritoneal graft mesothelium transformed into urothelium-like cells in the neobladder while having muscle distribution like that of the normal bladder. Gene set enrichment analysis revealed that neobladder was enriched in keratinization/epithelial cell differentiation, water homeostasis, and nucleobase containing compounds transporter activity, resembling the function of urothelium cells and not peritoneum cells. While pigs were able to urinate spontaneously after the catheter was removed post-surgically, innervation of the neobladder still remains a challenging problem. In this instance, the pig model has been vital in terms of tissue engineering research [91]. To our knowledge, bladder cancer animal-based translational research has largely been dominated by murine models. However, given the immense genetic similarities between swine and human compared to murine and human, along with the generation of the Oncopig model, an argument can be made regarding additional research for optimum human clinical translatability.
Interventions and potential for treatments using the pig model
There are plenty of available references for the use of swine in biomedical research. Pigs have been used to study transplantation, immunity pre- and postnatally, stress, allergies, transmittable diseases, vaccination, and cancer [92]. There has been considerable use of percutaneous thermal ablation by radiofrequency [41] or microwave [42, 43] based methods for the treatment of primary and metastatic hepatic malignancies using pig models. Pig models have been used to study the heat sink effect caused by vessels located close to the target area of ablation. This can result in incomplete tumor ablation and is therefore a known risk factor for local tumor recurrence post-procedure [43]. Liver tumors have been successfully induced in transgenic pigs and can also be successfully treated using transarterial embolization [44]. Newer therapeutic strategies such as histotripsy and focused ultrasound ablation with precise control of acoustic cavitation, have also been performed and studied with the use of pig models [45]. Biomedical technology companies, researchers, and physicians have used the porcine model to study viable options for minimally invasive treatment of primary and metastatic hepatic malignancies.
Cancer antigens have become a promising vaccine target and immunotherapy has played an overall role in survival for metastatic cancer patients. There exists large amount of cancer vaccination studies in mice; however, majority of these candidate vaccines have failed to provide a therapeutic response in subsequent human clinical trials. Indoleamine 2,3-dioxygenase (IDO) and Ras homolog gene family member C (RhoC) have been promising antigen targets for vaccine development against multiple cancer forms. Due to the advantage of recombinant swine MHC class I molecules (SLAs), cytotoxic T-cell inducing adjuvants were developed and the stability of peptide-SLA complex was assessed. Regardless of the adjuvant, the vaccine-induced peptide-specific cytotoxic T-cell responses were observed in both IDO and RhoC groups. However, there is more work to be done to develop a high throughput MHC multimer screening system for porcine cells, the pig model proves to be very translatable for optimum vaccine composition and formulation studies like number of injections for endogenous peptide immunization or their dosages [46]. One such example would be the humanized Gottingen minipigs carrying human genes for immunoglobulin heavy chains γ1 and γ4 and the immunoglobulin light chain κ [47]. When predicting patient responsiveness to immunotherapies, it is vital to consider the presence of intratumor immune cells as well as tumor microenvironment. KRASG12D and TP53R167H Oncopigs have opened a new paradigm to studying immunotherapies. Furthermore, antitumor immune responses have also been reported in Oncopigs. CD8β+ T cells were shown to specifically infiltrate Oncopig tumors, whereas the proportion of CD4+ T cells expressing CD8α+ activation molecule was significantly reduced in tumor. Compared to blood, a fourfold increase in γδ T cells displaying the CD2+CD8α+ T phenotype was observed in tumors. While the role of γδ T cells has not been proven definitively, there also exists a T-cell receptor delta constant (TRDC) region knockout pig which might shed more insight into this specific T-cell subpopulation [48].
Oncopigs have shown an active regulatory T-cell compartment with CD4-CD8α+FOXP3+ T cells in the circulating T-cell pool while FOXP3+ T cells were detected within the tumors. IDO1, cytotoxic T-lymphocyte-associated protein 4 (CTLA4), and programmed death-ligand 1 (PDL1), proteins that help malignant cancer cells escape T cell-mediated killing were significantly high in Oncopig leiomyosarcoma tumors. However, transformed cell lines from Oncopig (HCC and fibroblasts) showed no increases in expression of pro-survival proteins, meaning cellular transformation was likely not the cause of increased pro-survival protein expression in Oncopig leiomyosarcomas. Together these data showed that Oncopig immune system can recognize the AdCre-induced tumors and mount an antitumor immune response dominated by cytotoxic CD8β+ T cells, differentiated γδ T cells, with mediated regulatory response from FOXP3+ T cells and elevated expression of immunosuppressive genes. While there are some key immunologic differences between pigs and humans, pigs’ robust immune system still provides an important stage for studying and translating antitumor immune responses in humans [49].
Xenotransplantation of organs [50], tissues, and cells from genetically engineered pigs possesses immense therapeutic promise. Perhaps the most widely discussed recent story is of the first pig-to-human heart transplant [51]. Thus far efforts have been focused on overcoming the innate immune response; however, overcoming the adaptive immune response would be the next challenge. Currently administering anti-CD154 mAb, to block CD40/CD154 co-stimulation pathway, leads to suppressing the adaptive immune response. This enables the pig kidney graft, for example, to survive for many months without rejection. While there are many additional criteria to be considered for xenotransplantation in humans, pigs offer a real opportunity for the approximately 40% transplant candidates that have died, within 5 years, while waiting for an available organ [52]. Similar implications of xenotransplantation have been discussed with HCC or acute liver diseases with life-threatening liver dysfunction [53].
Lung cancer
Lung cancer is the second most diagnosed form of cancer after prostate cancer in males (12%) and breast cancer in females (13%). Lung and bronchus cancers, are however, the highest causes of cancer-related death in males and females (both 21%). While there have been advances in 3-year relative survival rate for all stages of lung cancer combined, reflecting towards early detection and advances in staging and surgical procedures, the death rate is still devastatingly high [2]. Animal models that can fulfill the gap in clinical translational research continue to be of importance in studying the pathophysiology of lung cancer. A major advance was made with the generation of the porcine cystic fibrosis model. To compare human and pig lung anatomy, neonatal porcine respiratory bronchioles are connected to paired alveolar ducts. In humans, respiratory bronchioles are connected to 2 - 3 primordial alveolar ducts at birth. The porcine trachea is also more cartilaginous and longer than the human trachea. Similar number of bronchial generations (including the size and bifurcations of the bronchial tree) has also been identified between pigs and humans. The porcine airway has a monopodial branching system in contrast to humans bipodial branching system. One especially distinguishing feature in pigs would be the right cranial lobe which arises from the right wall of the trachea before the bifurcation of the right and left main bronchus [93]. The presence of an independent right cranial lobe bronchus is extremely advantageous for performing interventions due to its ease of access. While both pigs and humans have highly lobulated lungs, in humans there is an incomplete collagenous component of interlobular septa, presence of interalveolar pores (of Kohn) and other communicating channels. In pigs, however, the collagenous component of the porcine interlobular septa is more complete meaning that collateral ventilation is less likely [93].
Due to the similarity between pig and human lungs, pigs have been extensively used for trialing minimally invasive procedures such as microwave ablation [54-56], understanding the differences in conventional versus cone beam CT-based monitoring techniques [57], and understanding xenogeneic cross-circulation to support human donor lung ex vivo [58, 59]. With the advancement of genetic modifications techniques and availability of whole pig genome sequencing data, genetic lesions in vivo can be recapitulated to understand tumor development. Single guide RNAs and CRISPR/Cas9-based system have been used to inactivate five tumor suppressor genes (TP53, PTEN, APC, BRCA1, and BRCA2) and one oncogene (KRAS) via lentiviral particles administered intranasally, resulting in rapid (3 months after infection) lung tumor development [60]. A transgenic pig model appears ideal to study lung cancer exposure, initiation/pathogenesis, and metastatic progression (Fig. 5). Very recently pulmonary nodules were induced in a transgenic pig model along with its preliminary characterization. AdCre was injected endovascularly into Oncopigs through the pulmonary arteries or the inferior vena cava. Whereas two other Oncopigs had a lung biopsy which was incubated with AdCre ex vivo before injecting the mixture into the lungs percutaneously. They observed one out of 10 endovascular inoculations and two out of six percutaneous inoculations (post lung biopsy and incubation with AdCre followed by percutaneous injection) resulted in neoplastic lung nodules [61]. Previously the same group was able to demonstrate a strong infiltrating CD8β+ predominance within the tumor microenvironment when compared to the peripheral T-cell pool [49]. Due to the presence of infiltrative CD8+ T cell in Oncopig tumors, the tumors regressed after 2 weeks. Additional research into lung cancer Oncopig models is warranted due to the high degree of translatability between pigs and humans. Differentiating between leaky CRISPR/Cas9 expression versus the presence of infiltrative CD8-positive T cells in Oncopig tumors and their regression is imperative to bridge the gap in current understanding of Oncopig antitumor immune responses. The Oncopig lung tumors express human adenocarcinoma markers (cytokeratin 7 and thyroid transcription factor I) but they appear to be histologically undifferentiated, meaning not showing glandular structures (adenocarcinoma), keratin pearls and intracellular bridges (squamous origin), neuroendocrine cells (small cell carcinoma), or pleomorphic giant cells (large cell carcinoma). Better Oncopig lung tumor characterization is needed to clarify the degree of clinical translatability. While it is promising that Oncopig tumor cells show progression through epithelial-mesenchymal transition, histopathological differentiation would lead to a superior translatable lung cancer model.
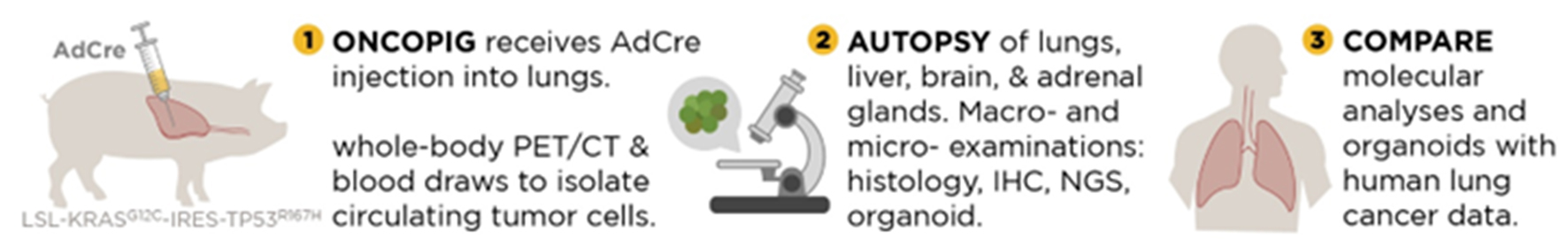 Click for large image | Figure 5. General schematic representation and workflow of Oncopig lung cancer model. |
| Conclusion With Translational Scope of Pig Models in General | ▴Top |
In conclusion, this review focused on using pigs as a preclinical cancer research model and summarized the advantages, disadvantages, and current models being used in cancer research (Fig. 1 and Table 1). Due to the full characterization of the pig genome and advancement of gene editing technologies, pigs have the potential to become a robust preclinical model for cancer research. High genetic synteny along with similarities in tissue-specific epigenetics between pigs and humans, versus mice and humans likely result in better translatability of findings. As cancer remains the second leading cause of death worldwide, it is imperative to develop a large animal model that offers optimum translatability to the human clinic. We have recapitulated how pigs were influential in brain, bone, blood, colon, pancreatic, liver, bladder, and lung cancer. Additionally, pigs have been used to develop and translate minimally invasive techniques to humans. There are limitations with the pig model such as requirement of more space, cost, time for development, and low availability of established models. Despite this, pigs can offer better translatability for human disease, and can aid in the education of the next generation of physicians. Therefore, we propose that pigs should become the new gold standard and gain popularity for translational medical research.
Acknowledgments
We thank the National Swine Resource and Research Center (NSRRC) at the University of Missouri for providing foundational Oncopigs for the research community. We thank Dr. Christine Elsik, University of Missouri, for her guidance with genetic and epigenetic analysis between humans, pigs, mouse, rats, and canines. We thank Samantha Peters, University of Missouri, for generating Figure 5. This study was funded in part by MU Mission Enhancement Funding, Department of Medicine, School of Medicine, University of Missouri-Columbia (TJH). J.T.K. and S.R. received pilot funding from the Ellis Fischel Cancer Center, University of Missouri.
Financial Disclosure
SR obtained funding from the National Institutes of Health and the National Cancer Institute R01 CA247763. TH, JK, SR: MU Mission Enhancement Funding, School of Medicine, University of Missouri-Columbia. BT, RSP; are supported in part by National Swine Resource and Research Center (NSRRC). Funding for the NSRRC is from the National Institute of Allergy and Infectious Disease, the National Institute of Heart, Lung and Blood, and the Office of Research Infrastructure Programs, Office of the Director (U42OD011140). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
BT is a founding member and serves as a consultant for RenOVAte Biosciences Inc. (RBI). All remaining authors declare no competing or conflict of interest.
Author Contributions
KJ conceived, edited and wrote the manuscript including the figures and tables, proof-read the manuscript and conducted analysis with finding gene orthologs in five species. TK aided in the development of the manuscript. AH and JC conducted analysis with finding gene orthologs in five species, proof-read and edited the manuscript. BT, RSP, KW, KEW, and JB proof-read and edited the manuscript. TH provided funding for KJ and edited the manuscript. JK was involved in the concept creation, drafted edited and proofread the manuscript. SR was involved in the concept creation, provided directions before, during and after the writing process, and has proof-read and edited the manuscript. All authors approved the final draft for submission.
Data Availability
The authors declare that all data generated or analyzed during this study are available within the article.
Abbreviations
AdCre: adenovirus expressing Cre recombinase; AFP: alpha-fetoprotein; ALS: amyotrophic lateral sclerosis; AML: acute myeloid leukemia; APC: adenomatous polyposis coli; ARID1A: AT-rich interactive domain-containing protein 1A; BCG: Bacillus Calmette-Guerin; CLL: chronic lymphocytic leukemia; CML: chronic myelogenous/myeloid leukemia; CRC: colorectal cancer; CRISPR/Cas9: clustered regularly interspaced short palindromic repeats/CRISPR-associated protein; CT: computed tomography; TRDC: T-cell receptor delta constant region; CTLA4: cytotoxic T-lymphocyte-associated protein 4; CyA: cyclosporin A; FAH: fumarylacetoacetate hydrolase; FAP: familial adenomatous polyposis; fl: floxed; GBM: glioblastoma or glioblastoma multiforme; G-CSF: granulocyte colony-stimulating factor; GE: genetic engineering; GEMM: genetically engineered mouse models; GFAP: glial fibrillary protein acids; GvHD: graft-versus-host disease; HCC: hepatocellular carcinoma; HCT: hematopoietic cell transplantation; HPD: 4-hydroxyphenylpyruvate dioxygenase; HT-1: hereditary tyrosinemia type-1; IDO: indoleamine 2,3-dioxygenase; IL: interleukin; ISC: intestinal epithelial stem cells; KPC: Pdx-Cre x LSL-KrasG12D-Trp53R172H; LDH: lactate dehydrogenase; LGR-5: leucine rich repeat containing G protein-coupled receptor 5; MGH-MHC: Massachusetts General Hospital major histocompatibility locus; MMP: matrix metalloproteinases; MSCs: mesenchymal stem cells; NK: natural killer cells; NTBC: 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione; Olfm4: olfactomedin-4; OS: osteosarcoma; PDAC: pancreatic ductal adenocarcinoma; Ph: Philadelphia chromosome; PNET: pancreatic neuroendocrine tumors; PTLD: post-transplantation lymphoproliferative disorder; PUB: porcine urinary bladder; RhoC: Ras homolog gene family member C; SCF: stem cell factor; SCID: severe combined immunodeficiency; SGCN: secretagogin; SLAs: MHC class molecules; SNP: single nucleotide polymorphisms; SV40: simian virus 40; SWI/SNF: SWItch/sucrose non-fermentable; TGF-β: transforming growth factor beta
| References | ▴Top |
- Xu J, Murphy SL, Kochanek KD. Mortality in the United States, 2021. NCHS data brief. 2022.
doi - Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17-48.
doi pubmed - Schachtschneider KM, Schwind RM, Newson J, Kinachtchouk N, Rizko M, Mendoza-Elias N, Grippo P, et al. The oncopig cancer model: an innovative large animal translational oncology platform. Front Oncol. 2017;7:190.
doi pubmed pmc - Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20(1):50-57.
doi pubmed pmc - Cheon DJ, Orsulic S. Mouse models of cancer. Annu Rev Pathol. 2011;6:95-119.
doi pubmed - Watson AL, Carlson DF, Largaespada DA, Hackett PB, Fahrenkrug SC. Engineered swine models of cancer. Front Genet. 2016;7:78.
doi pubmed pmc - Koscielny A. What is the value of animal models in laparoscopic surgery? A systematic review. Annals of Laparoscopic and Endoscopic Surgery. 2022;7:1.
- Sun J, Lu F, Luo Y, Bie L, Xu L, Wang Y. OrthoVenn3: an integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023;51(W1):W397-W403.
doi pubmed pmc - Bovine Genome S, Analysis C, Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324(5926):522-528.
doi pubmed pmc - De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269-1271.
doi pubmed - Thybert D, Roller M, Navarro FCP, Fiddes I, Streeter I, Feig C, Martin-Galvez D, et al. Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 2018;28(4):448-459.
doi pubmed pmc - Jorgensen FG, Hobolth A, Hornshoj H, Bendixen C, Fredholm M, Schierup MH. Comparative analysis of protein coding sequences from human, mouse and the domesticated pig. BMC Biol. 2005;3:2.
doi pubmed pmc - Pan Z, Yao Y, Yin H, Cai Z, Wang Y, Bai L, Kern C, et al. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat Commun. 2021;12(1):5848.
doi pubmed pmc - Dmochewitz M, Wolf E. Genetic engineering of pigs for the creation of translational models of human pathologies. Animal Frontiers. 2015;5(1):50-56.
- Lunney JK, Van Goor A, Walker KE, Hailstock T, Franklin J, Dai C. Importance of the pig as a human biomedical model. Sci Transl Med. 2021;13(621):eabd5758.
doi pubmed - Selek L, Seigneuret E, Nugue G, Wion D, Nissou MF, Salon C, Seurin MJ, et al. Imaging and histological characterization of a human brain xenograft in pig: the first induced glioma model in a large animal. J Neurosci Methods. 2014;221:159-165.
doi pubmed - Khoshnevis M, Carozzo C, Bonnefont-Rebeix C, Belluco S, Leveneur O, Chuzel T, Pillet-Michelland E, et al. Development of induced glioblastoma by implantation of a human xenograft in Yucatan minipig as a large animal model. J Neurosci Methods. 2017;282:61-68.
doi pubmed - Tora MS, Texakalidis P, Neill S, Wetzel J, Rindler RS, Hardcastle N, Nagarajan PP, et al. Lentiviral Vector Induced Modeling of High-Grade Spinal Cord Glioma in Minipigs. Sci Rep. 2020;10(1):5291.
doi pubmed pmc - Khoshnevis M, Carozzo C, Brown R, Bardies M, Bonnefont-Rebeix C, Belluco S, Nennig C, et al. Feasibility of intratumoral 165Holmium siloxane delivery to induced U87 glioblastoma in a large animal model, the Yucatan minipig. PLoS One. 2020;15(6):e0234772.
doi pubmed pmc - Sieren JC, Meyerholz DK, Wang XJ, Davis BT, Newell JD, Jr., Hammond E, Rohret JA, et al. Development and translational imaging of a TP53 porcine tumorigenesis model. J Clin Invest. 2014;124(9):4052-4066.
doi pubmed pmc - Saalfrank A, Janssen KP, Ravon M, Flisikowski K, Eser S, Steiger K, Flisikowska T, et al. A porcine model of osteosarcoma. Oncogenesis. 2016;5(3):e210.
doi pubmed pmc - Niu G, Hellmuth I, Flisikowska T, Pausch H, Rieblinger B, Carrapeiro A, Schade B, et al. Porcine model elucidates function of p53 isoform in carcinogenesis and reveals novel circTP53 RNA. Oncogene. 2021;40(10):1896-1908.
doi pubmed pmc - Lee PW, Cina RA, Randolph MA, Goodrich J, Rowland H, Arellano R, Kim HB, et al. Stable multilineage chimerism across full MHC barriers without graft-versus-host disease following in utero bone marrow transplantation in pigs. Exp Hematol. 2005;33(3):371-379.
doi pubmed - Powell EJ, Cunnick JE, Knetter SM, Loving CL, Waide EH, Dekkers JC, Tuggle CK. NK cells are intrinsically functional in pigs with Severe Combined Immunodeficiency (SCID) caused by spontaneous mutations in the Artemis gene. Vet Immunol Immunopathol. 2016;175:1-6.
doi pubmed pmc - Duran-Struuck R, Cho PS, Teague AG, Fishman B, Fishman AS, Hanekamp JS, Moran SG, et al. Myelogenous leukemia in adult inbred MHC-defined miniature swine: a model for human myeloid leukemias. Vet Immunol Immunopathol. 2010;135(3-4):243-256.
doi pubmed pmc - Matar AJ, Patil AR, Al-Musa A, Hanekamp I, Sachs DH, Huang CA, Duran-Struuck R. Effect of irradiation on incidence of post-transplant lymphoproliferative disorder after hematopoietic cell transplantation in miniature swine. Biol Blood Marrow Transplant. 2015;21(10):1732-1738.
doi pubmed - Boettcher AN, Li Y, Ahrens AP, Kiupel M, Byrne KA, Loving CL, Cino-Ozuna AG, et al. Novel engraftment and T cell differentiation of human hematopoietic Cells in ART(-/-)IL2RG(-/Y) SCID pigs. Front Immunol. 2020;11:100.
doi pubmed pmc - Winogrodzki T, Metwaly A, Grodziecki A, Liang W, Klinger B, Flisikowska T, Fischer K, et al. TNF DeltaARE pigs: a translational Crohn's disease model. J Crohns Colitis. 2023;17(7):1128-1138.
doi pubmed pmc - Flisikowska T, Merkl C, Landmann M, Eser S, Rezaei N, Cui X, Kurome M, et al. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143(5):1173-1175.e1177.
doi pubmed - Schaaf CR, Polkoff KM, Carter A, Stewart AS, Sheahan B, Freund J, Ginzel J, et al. A LGR5 reporter pig model closely resembles human intestine for improved study of stem cells in disease. FASEB J. 2023;37(6):e22975.
doi pubmed pmc - Principe DR, Overgaard NH, Park AJ, Diaz AM, Torres C, McKinney R, Dorman MJ, et al. KRAS(G12D) and TP53(R167H) cooperate to induce pancreatic ductal adenocarcinoma in sus scrofa pigs. Sci Rep. 2018;8(1):12548.
doi pubmed pmc - Berthelsen MF, Callesen MM, Ostergaard TS, Liu Y, Li R, Callesen H, Dagnaes-Hansen F, et al. Pancreas specific expression of oncogenes in a porcine model. Transgenic Res. 2017;26(5):603-612.
doi pubmed - Mondal P, Patel NS, Bailey K, Aravind S, Cartwright SB, Hollingsworth MA, Lazenby AJ, et al. Induction of pancreatic neoplasia in the KRAS/TP53 Oncopig. Dis Model Mech. 2023;16(1):1.
doi pubmed pmc - Schachtschneider KM, Schwind RM, Darfour-Oduro KA, De AK, Rund LA, Singh K, Principe DR, et al. A validated, transitional and translational porcine model of hepatocellular carcinoma. Oncotarget. 2017;8(38):63620-63634.
doi pubmed pmc - Gaba RC, Elkhadragy L, Boas FE, Chaki S, Chen HH, El-Kebir M, Garcia KD, et al. Development and comprehensive characterization of porcine hepatocellular carcinoma for translational liver cancer investigation. Oncotarget. 2020;11(28):2686-2701.
doi pubmed pmc - Yasmin A, Regan DP, Schook LB, Gaba RC, Schachtschneider KM. Transcriptional regulation of alcohol induced liver fibrosis in a translational porcine hepatocellular carcinoma model. Biochimie. 2021;182:73-84.
doi pubmed pmc - Elkhadragy L, Regan MR, W MT, Goli KD, Patel S, Garcia K, Stewart M, et al. Generation of genetically tailored porcine liver cancer cells by CRISPR/Cas9 editing. Biotechniques. 2021;70(1):37-48.
doi pubmed pmc - Elkhadragy L, Dasteh Goli K, Totura WM, Carlino MJ, Regan MR, Guzman G, Schook LB, et al. Effect of CRISPR knockout of AXIN1 or ARID1A on proliferation and migration of porcine hepatocellular carcinoma. Front Oncol. 2022;12:904031.
doi pubmed pmc - Nicolas CT, VanLith CJ, Hickey RD, Du Z, Hillin LG, Guthman RM, Cao WJ, et al. In vivo lentiviral vector gene therapy to cure hereditary tyrosinemia type 1 and prevent development of precancerous and cancerous lesions. Nat Commun. 2022;13(1):5012.
doi pubmed pmc - Boas FE, Nurili F, Bendet A, Cheleuitte-Nieves C, Basturk O, Askan G, Michel AO, et al. Induction and characterization of pancreatic cancer in a transgenic pig model. PLoS One. 2020;15(9):e0239391.
doi pubmed pmc - Poch FG, Rieder C, Ballhausen H, Knappe V, Ritz JP, Gemeinhardt O, Kreis ME, et al. The vascular cooling effect in hepatic multipolar radiofrequency ablation leads to incomplete ablation ex vivo. Int J Hyperthermia. 2016;32(7):749-756.
doi pubmed - Neizert CA, Do HNC, Zibell M, Rieder C, Sinden D, Niehues SM, Vahldiek JL, et al. Three-dimensional assessment of vascular cooling effects on hepatic microwave ablation in a standardized ex vivo model. Sci Rep. 2022;12(1):17061.
doi pubmed pmc - Ringe KI, Lutat C, Rieder C, Schenk A, Wacker F, Raatschen HJ. Experimental evaluation of the heat sink effect in hepatic microwave ablation. PLoS One. 2015;10(7):e0134301.
doi pubmed pmc - Nurili F, Monette S, Michel AO, Bendet A, Basturk O, Askan G, Cheleuitte-Nieves C, et al. Transarterial embolization of liver cancer in a transgenic pig model. J Vasc Interv Radiol. 2021;32(4):510-517.e513.
doi pubmed pmc - Hendricks-Wenger A, Arnold L, Gannon J, Simon A, Singh N, Sheppard H, Nagai-Singer MA, et al. Histotripsy ablation in preclinical animal models of cancer and spontaneous tumors in veterinary patients: a review. IEEE Trans Ultrason Ferroelectr Freq Control. 2022;69(1):5-26.
doi pubmed pmc - Overgaard NH, Frosig TM, Welner S, Rasmussen M, Ilsoe M, Sorensen MR, Andersen MH, et al. Establishing the pig as a large animal model for vaccine development against human cancer. Front Genet. 2015;6:286.
doi pubmed pmc - Flisikowska T, Egli J, Flisikowski K, Stumbaum M, Kung E, Ebeling M, Schmucki R, et al. A humanized minipig model for the toxicological testing of therapeutic recombinant antibodies. Nat Biomed Eng. 2022;6(11):1248-1256.
doi pubmed pmc - Petersen B, Kammerer R, Frenzel A, Hassel P, Dau TH, Becker R, Breithaupt A, et al. Generation and first characterization of TRDC-knockout pigs lacking gammadelta T cells. Sci Rep. 2021;11(1):14965.
doi pubmed pmc - Overgaard NH, Principe DR, Schachtschneider KM, Jakobsen JT, Rund LA, Grippo PJ, Schook LB, et al. Genetically induced tumors in the oncopig model invoke an antitumor immune response dominated by cytotoxic CD8beta(+) T cells and differentiated gammadelta T cells alongside a regulatory response mediated by FOXP3(+) T cells and immunoregulatory molecules. Front Immunol. 2018;9:1301.
doi pubmed pmc - Montgomery RA, Stern JM, Lonze BE, Tatapudi VS, Mangiola M, Wu M, Weldon E, et al. Results of two cases of pig-to-human kidney xenotransplantation. N Engl J Med. 2022;386(20):1889-1898.
doi pubmed - Madhusoodanan J. After the first pig-to-human heart transplant, scientists look to the future of cardiac xenotransplantation. JAMA. 2022;328(20):1999-2001.
doi pubmed - Cooper DKC, Hara H, Iwase H, Yamamoto T, Jagdale A, Kumar V, Mannon RB, et al. Clinical pig kidney xenotransplantation: how close are we? J Am Soc Nephrol. 2020;31(1):12-21.
doi pubmed pmc - Li X, Wang Y, Yang H, Dai Y. Liver and hepatocyte transplantation: what can pigs contribute? Front Immunol. 2021;12:802692.
doi pubmed pmc - De Leon H, Royalty K, Mingione L, Jaekel D, Periyasamy S, Wilson D, Laeseke P, et al. Device safety assessment of bronchoscopic microwave ablation of normal swine peripheral lung using robotic-assisted bronchoscopy. Int J Hyperthermia. 2023;40(1):2187743.
doi pubmed - Ghosn M, Elsakka AS, Ridouani F, Doustaly R, Mingione L, Royalty K, Ziv E, et al. Augmented fluoroscopy guided transbronchial pulmonary microwave ablation using a steerable sheath. Transl Lung Cancer Res. 2022;11(2):150-164.
doi pubmed pmc - Kodama H, Ueshima E, Gao S, Monette S, Paluch LR, Howk K, Erinjeri JP, et al. High power microwave ablation of normal swine lung: impact of duration of energy delivery on adverse event and heat sink effects. Int J Hyperthermia. 2018;34(8):1186-1193.
doi pubmed pmc - Meram E, Longhurst C, Brace CL, Laeseke PF. Comparison of conventional and cone-beam CT for monitoring and assessing pulmonary microwave ablation in a porcine model. J Vasc Interv Radiol. 2018;29(10):1447-1454.
doi pubmed - Wu WK, Stier MT, Stokes JW, Ukita R, Patel YJ, Cortelli M, Landstreet SR, et al. Immune characterization of a xenogeneic human lung cross-circulation support system. Sci Adv. 2023;9(13):eade7647.
doi pubmed pmc - Kelly Wu W, Guenthart BA, O'Neill JD, Hozain AE, Tipograf Y, Ukita R, Stokes JW, et al. Technique for xenogeneic cross-circulation to support human donor lungs ex vivo. J Heart Lung Transplant. 2023;42(3):335-344.
doi pubmed pmc - Wang K, Jin Q, Ruan D, Yang Y, Liu Q, Wu H, Zhou Z, et al. Cre-dependent Cas9-expressing pigs enable efficient in vivo genome editing. Genome Res. 2017;27(12):2061-2071.
doi pubmed pmc - Ghosn M, Elsakka AS, Petre EN, Cheleuitte-Nieves C, Tammela T, Monette S, Ziv E, et al. Induction and preliminary characterization of neoplastic pulmonary nodules in a transgenic pig model. Lung Cancer. 2023;178:157-165.
doi pubmed pmc - Leuchs S, Saalfrank A, Merkl C, Flisikowska T, Edlinger M, Durkovic M, Rezaei N, et al. Inactivation and inducible oncogenic mutation of p53 in gene targeted pigs. PLoS One. 2012;7(10):e43323.
doi pubmed pmc - DeFelipe J. The anatomical problem posed by brain complexity and size: a potential solution. Front Neuroanat. 2015;9:104.
doi pubmed pmc - Sauleau P, Lapouble E, Val-Laillet D, Malbert CH. The pig model in brain imaging and neurosurgery. Animal. 2009;3(8):1138-1151.
doi pubmed - Ganne A, Balasubramaniam M, Griffin WST, Shmookler Reis RJ, Ayyadevara S. Glial fibrillary acidic protein: a biomarker and drug target for Alzheimer's disease. Pharmaceutics. 2022;14(7):1354.
doi pubmed pmc - Gregory JM, Elliott E, McDade K, Bak T, Pal S, Chandran S, Abrahams S, et al. Neuronal clusterin expression is associated with cognitive protection in amyotrophic lateral sclerosis. Neuropathol Appl Neurobiol. 2020;46(3):255-263.
doi pubmed pmc - Sjostedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, Oksvold P, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367(6482):eaay5947.
doi pubmed - Hicks WH, Bird CE, Pernik MN, Haider AS, Dobariya A, Abdullah KG, Aoun SG, et al. Large animal models of glioma: current status and future prospects. Anticancer Res. 2021;41(11):5343-5353.
doi pubmed - Sansone G, Pini L, Salvalaggio A, Gaiola M, Volpin F, Baro V, Padovan M, et al. Patterns of gray and white matter functional networks involvement in glioblastoma patients: indirect mapping from clinical MRI scans. Front Neurol. 2023;14:1175576.
doi pubmed pmc - Jarvis S, Koumadoraki E, Madouros N, Sharif S, Saleem A, Khan S. Non-rodent animal models of osteosarcoma: A review. Cancer Treat Res Commun. 2021;27:100307.
doi pubmed - Takeda A, Yaseen NR. Nucleoporins and nucleocytoplasmic transport in hematologic malignancies. Semin Cancer Biol. 2014;27:3-10.
doi pubmed - Duran-Struuck R, Huang CA, Matar AJ. Cellular therapies for the treatment of hematological malignancies; Swine are an ideal preclinical model. Front Oncol. 2019;9:418.
doi pubmed pmc - AbuSamra DB, Aleisa FA, Al-Amoodi AS, Jalal Ahmed HM, Chin CJ, Abuelela AF, Bergam P, et al. Not just a marker: CD34 on human hematopoietic stem/progenitor cells dominates vascular selectin binding along with CD44. Blood Adv. 2017;1(27):2799-2816.
doi pubmed pmc - Ye N, Cai J, Dong Y, Chen H, Bo Z, Zhao X, Xia M, et al. A multi-omic approach reveals utility of CD45 expression in prognosis and novel target discovery. Front Genet. 2022;13:928328.
doi pubmed pmc - Schaaf CR, Gonzalez LM. Use of translational, genetically modified porcine models to ultimately improve intestinal disease treatment. Front Vet Sci. 2022;9:878952.
doi pubmed pmc - Li WC, Zhang HM, Li J, Dong RK, Yao BC, He XJ, Wang HQ, et al. Comparison of biomechanical properties of bile duct between pigs and humans for liver xenotransplant. Transplant Proc. 2013;45(2):741-747.
doi pubmed - Charepalli V, Reddivari L, Radhakrishnan S, Eriksson E, Xiao X, Kim SW, Shen F, et al. Pigs, Unlike mice, have two distinct colonic stem cell populations similar to humans that respond to high-calorie diet prior to insulin resistance. Cancer Prev Res (Phila). 2017;10(8):442-450.
doi pubmed pmc - Flisikowski K, Perleberg C, Niu G, Winogrodzki T, Bak A, Liang W, Grodziecki A, et al. Wild-type APC influences the severity of familial adenomatous polyposis. Cell Mol Gastroenterol Hepatol. 2022;13(2):669-671.e663.
doi pubmed pmc - Yim JJ, Harmsen S, Flisikowski K, Flisikowska T, Namkoong H, Garland M, van den Berg NS, et al. A protease-activated, near-infrared fluorescent probe for early endoscopic detection of premalignant gastrointestinal lesions. Proc Natl Acad Sci U S A. 2021;118(1):e2008072118.
doi pubmed pmc - Rogalla S, Flisikowski K, Gorpas D, Mayer AT, Flisikowska T, Mandella MJ, Ma X, et al. Biodegradable fluorescent nanoparticles for endoscopic detection of colorectal carcinogenesis. Adv Funct Mater. 2019;29(51):1904992.
doi pubmed pmc - Troya J, Krenzer A, Flisikowski K, Sudarevic B, Banck M, Hann A, Puppe F, et al. New concept for colonoscopy including side optics and artificial intelligence. Gastrointest Endosc. 2022;95(4):794-798.
doi pubmed - Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7-33.
doi pubmed - Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1-2):349-361.
doi pubmed pmc - Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469-483.
doi pubmed - Bailey KL, Cartwright SB, Patel NS, Remmers N, Lazenby AJ, Hollingsworth MA, Carlson MA. Porcine pancreatic ductal epithelial cells transformed with KRAS(G12D) and SV40T are tumorigenic. Sci Rep. 2021;11(1):13436.
doi pubmed pmc - Melzer MK, Breunig M, Arnold F, Wezel F, Azoitei A, Roger E, Kruger J, et al. Organoids at the PUB: the porcine urinary bladder serves as a pancreatic niche for advanced cancer modeling. Adv Healthc Mater. 2022;11(11):e2102345.
doi pubmed - Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873(1):188314.
doi pubmed pmc - John BA, Said N. Insights from animal models of bladder cancer: recent advances, challenges, and opportunities. Oncotarget. 2017;8(34):57766-57781.
doi pubmed pmc - Zhang N, Li D, Shao J, Wang X. Animal models for bladder cancer: The model establishment and evaluation (Review). Oncol Lett. 2015;9(4):1515-1519.
doi pubmed pmc - Baust JM, et al. Evaluation of a novel cystoscopic compatible cryocatheter for the treatment of Bladder Cancer. Bladder Cancer. 2020;6:303-318.
- Chen B, Chen X, Wang W, Shen J, Song Z, Ji H, Zhang F, et al. Tissue-engineered autologous peritoneal grafts for bladder reconstruction in a porcine model. J Tissue Eng. 2021;12:2041731420986796.
doi pubmed pmc - Kaser T. Swine as biomedical animal model for T-cell research-Success and potential for transmittable and non-transmittable human diseases. Mol Immunol. 2021;135:95-115.
doi pubmed - Judge EP, Hughes JM, Egan JJ, Maguire M, Molloy EL, O'Dea S. Anatomy and bronchoscopy of the porcine lung. A model for translational respiratory medicine. Am J Respir Cell Mol Biol. 2014;51(3):334-343.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


