| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 4, August 2024, pages 612-624
High Probability of Lynch Syndrome Among Colorectal Cancer Patients Is Associated With Higher Occurrence of KRAS and PIK3CA Mutations
Didik Setyo Heriyantoa, b , Naomi Yoshuantaria
, Gilang Akbarianic
, Vincent Laua
, Hanifa Haninic
, Zulfa Hidayatic
, Muhammad Zulfikar Ariefc
, Andrew Nobiantoro Gunawana
, Asep Muhamad Ridwanulohd
, Wien Kusharyotod
, Adeodatus Yuda Handayae
, Mohammad Ilyasf
, Johan Kurniandag
, Susanna Hilda Hutajulug
, Susanti Susantic, f, h, i
aDepartment of Anatomical Pathology, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada/Dr Sardjito General Hospital Yogyakarta, Indonesia
bCollaboration Research Center for Precision Oncology based Omics - PKR PrOmics, Yogyakarta, Indonesia
cPathgen Diagnostik Teknologi, Ir. Soekarno Science and Technology Park, National Research and Innovation Agency Republic of Indonesia, Bogor, Indonesia
dResearch Center for Genetic Engineering, Research Organization for Life Sciences and Environment, National Research and Innovation Agency Republic of Indonesia, Ir. Soekarno Science and Technology Park, Bogor, Indonesia
eDivision of Digestive Surgeon, Department of Surgery, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada/Dr Sardjito General Hospital Yogyakarta, Indonesia
fMolecular Pathology Research Group, Academic Unit of Translational Medical Science, Biodiscovery Institute, School of Medicine, University of Nottingham, Nottingham, UK
gDivision of Hematology and Medical Oncology, Department of Internal Medicine, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada/Dr Sardjito General Hospital, Yogyakarta, Indonesia
hDepartment of Pharmacology and Clinical Pharmacy, Faculty of Pharmacy, Universitas Muhammadiyah Purwokerto, Indonesia
iCorresponding Author: Susanti Susanti, Pathgen Diagnostik Teknologi, Ir. Soekarno Science and Technology Park, National Research and Innovation Agency Republic of Indonesia, Bogor 16911, Indonesia
Manuscript submitted February 23, 2024, accepted May 25, 2024, published online June 17, 2024
Short title: KRAS/PIK3CA Mutations and Lynch Syndrome CRC
doi: https://doi.org/10.14740/wjon1843
| Abstract | ▴Top |
Background: In Indonesia, early-onset colorectal cancer (EOCRC) rates are higher in patients < 50 years old compared to Western populations, possibly due to a higher frequency of Lynch syndrome (LS) in CRC patients. We aimed to examine the association of KRAS and PIK3CA mutations with LS.
Methods: In this retrospective cross-sectional single-center study, the PCR-HRM-based test was used for screening of microsatellite instability (MSI) mononucleotide markers (BAT25, BAT26, BCAT25, MYB, EWSR1), MLH1 promoter methylation, and oncogene mutations of BRAF (V600E), KRAS (exon 2 and 3), and PIK3CA (exon 9 and 20) in FFPE DNA samples.
Results: All the samples (n = 244) were from Dr. Sardjito General Hospital Yogyakarta, Indonesia. KRAS and PIK3CA mutations were found in 151/244 (61.88%) and 107/244 (43.85%) of samples, respectively. KRAS and PIK3CA mutations were significantly associated with MSI status in 32/42 (76.19%) and 25/42 (59.52%) of samples, respectively. KRAS mutation was significantly associated with LS status in 26/32 (81.25%) of samples. The PIK3CA mutation was present in a higher proportion in LS samples of 19/32 (59.38%), but not statistically significant. Clinicopathology showed that KRAS mutation was significantly associated with right-sided CRC and higher histology grade in 39/151 (25.83%) and 24/151 (16.44%) samples, respectively. PIK3CA mutation was significantly associated with female sex and lower levels of tumor-infiltrating lymphocytes in 62/107 (57.94%) and 26/107 (30.23%) samples, respectively. KRAS and PIK3CA mutations did not significantly affect overall survival (120 months) in LS and non-LS patients.
Conclusions: The high probability of LS in Indonesian CRC patients is associated with KRAS and PIK3CA mutations.
Keywords: Colorectal neoplasms; Pathology; Molecular; Medical oncology; Gastrointestinal neoplasms; Neoplastic syndromes; Hereditary
| Introduction | ▴Top |
Colorectal cancer (CRC) is the third most prevalent cancer worldwide and as well as one of the deadliest. Approximately in Indonesia, over 35,000 patients are diagnosed with CRC each year [1]. Three provinces in Indonesia have the highest incidence of CRC: Jakarta, Central Java, and Yogyakarta. Early-onset colorectal cancer (EOCRC) accounts for nearly 30% of total CRC patients, three times higher than in Europe, the UK, and the USA [2]. The epidemiological data in Indonesia showed that the proportion of CRC patients < 40 years old was more than 30% [3]. This incidence in Indonesia was higher in males (54%) than in females (46%), with a peak age of 50 - 54 years [2]. Lynch syndrome (LS), also known as hereditary non-polyposis colon cancer (HNPCC) is a hereditary type of CRC. This syndrome is characterized by early-onset (< 50 years) [4, 5]. Our previous study introduced the higher frequency of LS cases in Yogyakarta, Indonesia linked to a high risk of EOCRC [6].
Three molecular pathways have been identified for the pathogenesis of CRC: chromosomal instability (CIN), microsatellite instability (MSI), and CpG island methylator phenotype (CIMP) [7, 8]. The CIN involves gene mutations in APC, KRAS, SMAD4, and TP53, while MSI is caused by mutations in mismatch repair (MMR) genes such as MLH1, MSH2, MSH6, and PMS2 [9]. The CIMP, which is characterized by CG dinucleotide methylation in the promoters of numerous genes, is associated with distinct clinical and pathological attributes in tumors. These subtypes, as described by Jass classification, include CIMP high/MSI high (12% of CRC), CIMP low/MSI low or microsatellite stable (20%), CIMP negative/microsatellite stable (57%), and HNPCC, with CIMP negative/MSI high and negative for BRAF mutations [7, 8, 10-12]. Testing for MSI and MMR protein deficiency (dMMR) is commonly the first step in LS diagnostics due to germline mutations of MMR genes. On the contrary, epigenetic silencing of the MLH1 and somatic mutation of BRAF are common in sporadic tumors with MSI but very rarely occur in tumors arising in LS [13, 14]. Approximately 10-15% of sporadic CRCs will also show dMMR/MSI due to somatic loss of MMR function [15]. Epigenetic silencing of the MLH1 gene is the most common cause of dMMR in sporadic tumors and very rarely occurs in LS. Thus, sporadic tumors with dMMR/MSI can be distinguished from tumors arising in LS by demonstrating methylation of the MLH1 promoter [16].
KRAS mutations, the most common RAS family mutation, affect cell proliferation, differentiation, senescence, and apoptosis in 40% of sporadic CRC. These mutations increase CRC tumor aggressiveness, reduce survival rates, and promote treatment resistance [17]. Anti-epidermal growth factor receptor (EGFR) monoclonal antibodies, such as cetuximab and panitumumab, are ineffective in CRC patients with KRAS codon 12, 13, or 14 mutations. Hence, these agents are only effective in RAS wild-type tumors [18]. BRAF mediates RAS-RAF-MAP kinase growth signal responses. BRAF mutations are found in 4% of MSI-low tumors and 40% of MSI-high tumors [19]. The most frequent of these mutations are BRAFV600E (Val600Glu). These BRAFV600E mutations help to differentiate between familial and sporadic CRC and are associated with poorer prognosis. Generally, BRAF mutations are confined to tumors without KRAS exon 2 mutations. BRAF is downstream of activated KRAS in the EGFR pathway, making cetuximab or panitumumab ineffective for inhibiting EGFR, unless given BRAF inhibitor [10, 20].
Phosphatidylinositol-3-kinase (PI3K) is a heterodimeric lipid kinase involved in cell signaling and cell membrane function. Mutations in PIK3CA have been reported in 10-20% of CRC cases [21]. PIK3CA mutations are associated with worse clinical outcomes and a negative predictor of response to anti-EGFR targeted therapy [22]. It has been shown that RAS mutations are negative predictors of anti-EGFR mAb response and survival benefit [22]. Over 80% of PIK3CA mutations are found in two hotspots: the helicase domain of exon 9 and the kinase domain in exon 20. Several studies have analyzed PIK3CA mutation in these hotspots for the discrepancy of the predictive values of PIK3CA as a biomarker for anti-EGFR [22, 23]. The PI3K pathway’s downstream effectors include AKT and mTOR, which increase cell cycle regulator mRNA translation [24]. The decreased expression of tumor suppressor gene PTEN, a direct antagonist, has been shown to be correlated with poor outcomes in CRC [25].
The possibility of LS can be inferred if a tumor is shown to be dMMR or shown to have MSI, but a definitive diagnosis of LS can only be made by demonstration of a germline mutation in MMR gene. KRAS mutations tend to occur in the context of CIN, which is characteristic of MSS tumors [26, 27]. Meanwhile, PIK3CA mutations have been found to be more prevalent in MSI tumors [28]. In Indonesia, there has been limited investigation into the genetic mutation profiles of CRC patients, particularly those with LS. As reported in our previous study and others, several clinical features are associated with this syndrome, including tumor location of which 60-70% were found in the right-sided (proximal) colon [6, 10]. In this study, we examine the association of oncogenic mutations of KRAS and PIK3CA with MSI, MLH1 promoter methylation and LS as well as the demography and clinicopathology profile of CRC patients in Yogyakarta, Indonesia.
| Materials and Methods | ▴Top |
Ethical statements
This study was approved by the Medical and Health Research Ethics Committee (MHREC) Faculty of Medicine, Public Health, and Nursing of Universitas Gadjah Mada, Yogyakarta (Ethical Approval Number KE/FK/0837/EC/2022). Informed consent has been obtained for the use of tumor samples, clinical data and any other relevant data in the research for all subjects.
CRC clinical samples
For this observational cross-sectional study, we performed retrospective consecutive sampling. A total of 288 formalin-fixed paraffin-embedded (FFPE) CRC samples from the primary tumors, with no data about simultaneous or metachronous metastasis, were collected from the Department of Anatomical Pathology at Dr. Sardjito General Hospital Yogyakarta, Indonesia between 2016 and 2021. The samples were limited due to the hospital being the national tertiary referral hospital, where many of the patients received treatment in our center (Dr. Sardjito General Hospital) following resection procedures that had been conducted in secondary/regional hospitals elsewhere. Of these, 244 CRC samples were eligible for mutation detection. The patient data acquired for each case included sociodemographic (age and sex), tumor pathology (location/site, staging analysis by TNM staging system, histologic grade, lymphovascular invasion status, morphology, tumor-infiltrating lymphocytes (TILs), and various clinical parameters (hemoglobin (Hb), Eastern Cooperative Oncology Group (ECOG) scale, and body mass index (BMI)).
DNA extraction
Paraffin blocks from CRC patients were cut into six pieces with 5 µm thickness. Only one piece in one slide continued with DNA extraction. Genomic DNA was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, USA) according to the manufacturer’s protocol. DNA samples were quantified using a NanoDrop™ spectrophotometer (Thermo Scientific, Waltham, MA, USA). Samples of sufficient concentration and quality were adjusted to a concentration of 20 ng/µL for PCR applications.
Detection of MSI, BRAF, MLH1 and oncogenes mutation
All CRC biomarkers were detected using an IVD kit called BioColomelt-Dx manufactured by Biofarma Ltd, Indonesia. BioColomelt-Dx is a PCR-HRM molecular diagnostics kit for screening MSI, MLH1 promoter methylation, and important oncogene mutations of KRAS (exon 2 and 3), BRAF (V600E), PIK3CA (exon 9 and 20) in FFPE DNA samples. MSI, BRAF, and MLH1 methylation promoter detections were previously described as N-LyST panel [29]. The N_LyST panel is a detection method for five mononucleotide microsatellite repeats, BRAFV600E mutations, and MLH1 region C promoter methylation status. For MSI analysis, samples were regarded as MSI if > 2 markers (40%) showed instability; otherwise, they were regarded as MSS tumors. Samples showing MSI, BRAF wildtype, and MLH1 promoter methylation (unmethylated) were classified as “probable Lynch”. Out of the total samples, 244 were successfully determined for oncogene mutation detection, while 223 were suitable for probable Lynch determination.
Statistical analysis
Correlation between variables was calculated using two-sided Fisher’s exact test; the test is considered significant if the P-value < 0.05. The overall survival analysis was conducted using a log-rank test to compare between groups and visualized with Kaplan-Meier curves. All analyses were performed using R software version 4.3.2.
| Results | ▴Top |
Association between KRAS and PIK3CA mutation with MSI status
The analysis of KRAS and PIK3CA with MSI status is shown in Table 1. There was a significant association between mutant KRAS and MSI vs. MSS (76.19% vs. 58.91%; P-value = 0.038); and mutant PIK3CA with MSI vs. MSS (59.52% vs. 40.59%; P-value = 0.027). There were 14/244 samples that had the concomitant mutation of KRAS and BRAF. There was no significant association between KRAS and BRAF concomitant mutation with MSI status (7.14% vs. 5.45%; P-value = 0.7).
 Click to view | Table 1. KRAS and PIK3CA With MSI Mutational Status |
Association between KRAS and PIK3CA mutation with probable Lynch status
The association of KRAS, PIK3CA and Lynch status is shown in Table 2. There was a significant association between mutant KRAS and probable Lynch status (81.25% vs. 58.64%; P-value = 0.018). However, there was no significant association between PIK3CA and probable Lynch status.
 Click to view | Table 2. KRAS, PIK3CA and Probable Lynch Status |
Clinicopathology association with oncogene status
As shown in Table 3, PIK3CA gene mutation frequency was higher in female patients compared to the male patients (57.94% vs. 42.06%; P-value = 0.040). The lower level of TILs was found in mutant PIK3CA (30.23% vs. 16.24%; P-value = 0.021). The mutation rate of KRAS in the right sided was higher and statistically significant than the left sided (25.83% vs. 13.04%; P-value = 0.022). Mutant KRAS was significantly higher in histology grade 3 compared to wild-type KRAS (16.44% vs. 7.53%; P-value = 0.038). There was no significant association between other clinicopathology parameters with KRAS oncogene status.
 Click to view | Table 3. Clinicopathology Association With PIK3CA and KRAS Oncogene Status |
Overall survival on KRAS and PIK3CA mutation stratified by LS status
We analyzed the overall survival based on KRAS and PIK3CA mutations stratified by LS status. The numbers of samples that met the criteria for this analysis on PIK3CA and KRAS mutation are 220 and 222, respectively. There were no statistically significant differences of overall survival (follow-up period of 120 month) based on KRAS and PIK3CA mutation with probable Lynch and non-Lynch status patient (Fig. 1). Additionally, we conducted an overall survival analysis considering PIK3CA and KRAS mutations stratified by MSI status (Supplementary Material 1, www.wjon.org), comparing PIK3CA and KRAS mutant versus wild-type cases (Supplementary Material 2, www.wjon.org), and differentiating probable Lynch from sporadic cases (Supplementary Material 3, www.wjon.org). None of these analyses yielded statistically significant results.
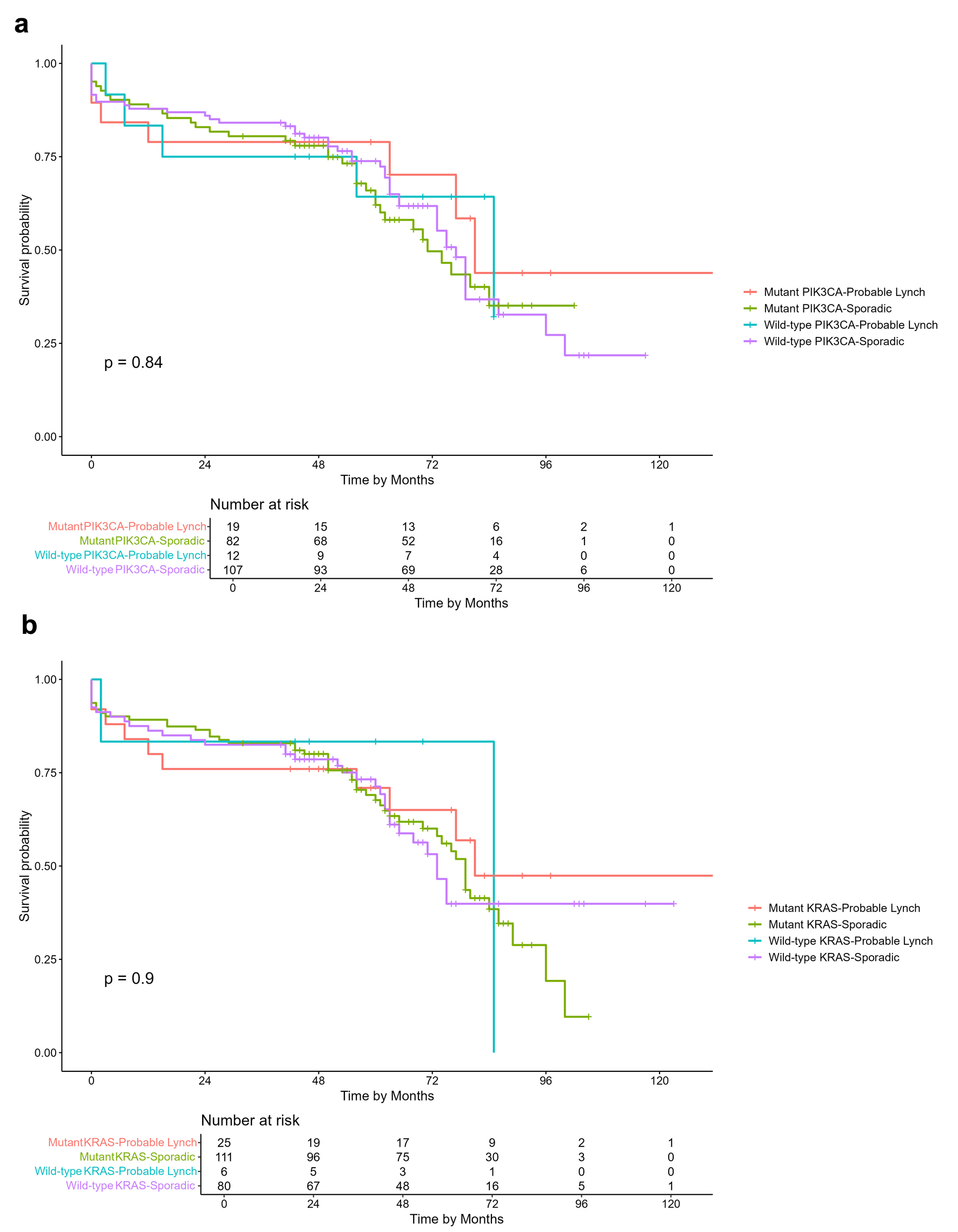 Click for large image | Figure 1. Kaplan-Meier overall survival curves of CRC patients. (a) Overall survival comparison based on PIK3CA mutation and probable Lynch syndrome status. (b) Overall survival comparison based on KRAS mutation and probable Lynch syndrome status. |
| Discussion | ▴Top |
As molecular testing for CRC is not routinely performed in clinical settings, currently there are only limited data on molecular landscape of CRC among the Indonesian patients. We previously reported higher incidence of EOCRC and probable Lynch in our patient cohort from Yogyakarta, Indonesia [6]. In this study, we sought to examine the association of oncogenic mutations of KRAS and PIK3CA, LS status and the clinicopathological features. As a brief description, Indonesia has a total population of approximately 238 million, with 131.48 million females compared to 132.68 million males [30, 31]. The country has a predominantly young age population, particularly from 20 to 40 years old [32]. The agricultural sector plays a significant role in socioeconomic development [33]. Yogyakarta is one of the most populated provinces in Indonesia. As of 2023, the total population accounted for 3.7 million people, mainly between 20 and 24 years of age [34]. The sex ratio between male and female population is 0.98. Yogyakarta is the region with the highest incidence of cancer in Indonesia, hence, highlighting the importance of this study [35].
The frequency of KRAS mutations (exon 2 and exon 3) in our cohort reached 61.88%. This was much higher than what was reported in the Asia-Pacific (37-52%) and Western populations (32-49%) [36-49]. Nevertheless, our data are in line with a previous study in Indonesia, showing that KRAS mutation was found in 71.8% of serrated adenocarcinoma (SA), which is intriguingly higher than the generally reported incidence of 40% [50-52]. Another study, based on next-generation sequencing (NGS) analysis, showed that KRAS mutation occurred in 63.6% of 22 Indonesian patients with mostly advanced CRC [53]. Distinct mutations located in codons 13, 14, 34, 58, 59, and 146 were found as opposed to more commonly reported mutations in codons 12, 13, 61, 146 [53-55]. A recent study in China has also reported that KRAS mutation was found in 69.4% of the early lesions [56, 57].
Furthermore, in this study, we found that the KRAS mutation was enriched in MSI compared to MSS cases (76.19% and 58.91%, respectively). We previously reported a lower frequency of BRAF mutations (20.45%) in this MSI-CRC cohort [6], similar to what has been reported in China [58-60]. This is unlike a common dogma in which KRAS mutation is more associated with MSS while BRAF mutation is associated with MSI [61-63]. The frequency of KRAS mutations in MSI-H CRCs has been reported to be approximately 12-38% [64-68]. This conflicting finding is likely due to the variability in the frequency of specific mutations in KRAS codons 12 and 13. A study by Asaka et al (2009) investigated KRAS mutations in different MSI status (MSI-H, MSI-L, and MSS) and reported that 93% G to A KRAS mutation occurred in MSI-H tumors compared to MSI-L and MSS [69]. Although we did not specifically examine this hotspot mutation, this phenomenon likely explains our findings.
We also observed the concomitant KRAS and BRAF mutation on 5.73% of total CRC cases. Despite previously thought as a rare event, there are a growing number of studies reporting the co-mutation, including a study by Gong et al (2017) showing the incidence rate of 1.4% of 138 metastatic CRC [70]. Our cohort was highly enriched for metastatic disease (41.1%) which may contribute to the higher frequency of co-mutation. However, it is important to note that most patients of our cohort presented with advanced disease status. To date, there is no optimal treatment for metastatic CRC harboring both KRAS and BRAF [71]. Consequently, our high co-mutation rate in the Indonesian population can shed new light that warrants further investigation. This study and others have shown that KRAS mutation was more frequent in right-sided CRC [48, 72, 73]. The predilection of KRAS mutations for the right side of the colon may be influenced by the fact that the right and left sides of the colon have different biology and histopathology in their respective embryological origins [48, 74, 75]. Right-sided CRC often has flat histopathology and a DNA MMR pathway deficiency [76]. This may explain the significant association of mutant KRAS and MSI in our cohort.
Our study reports the PIK3CA (exon 9 and 20) mutation frequency of 43.85%, which is lower compared to a previous study in other regions of Indonesia with smaller sample size, reporting 70.9% mutation [77]. An NGS-based study on Indonesian patients with advanced CRC revealed that all patients (100%, n = 22) harbor PIK3CA mutation, distinctively in exon 2, 5, 7, 8, 10, 19, and 21, in addition to commonly reported exon 9 and 20 [53]. Similar to our result, a meta-analysis of 44 studies enrolling 17,621 patients has reported that mutations of PIK3CA exon 9 and 20 were associated with MSI, KRAS mutation and right-sided colon [78]. Tumor with MSI is usually associated with a high number of TILs due to the generation of neoantigens, hence it serves as a good candidate for immunotherapy [79-81]. However, in this study, we found a subset of cases with lower numbers of TILs in PIK3CA mutant tumors, suggesting an immune evasion mechanism at play.
Interestingly, programmed death-ligand 1 (PD-L1) expression was correlated with PIK3CA mutations, suggesting that cancers with PIK3CA mutations and PD-L1 expression are immunotherapy candidates [28]. Inhibition of the PI3K-AKT pathway may improve effector T-cell infiltration in PI3K-altered CRC. Combining PI3K inhibitors with anti-PD-1 could enhance treatment efficacy and CD8+ T cell proliferation [82]. It is also worth noting that PIK3CA mutations are prevalent in the early stages of MSI, while it tends to occur later in MSS [83]. These suggest that the combination of PIK3CA mutation, MSI status, and PD-L1 expression could potentially be used to guide treatment selection or prognosis improvement for certain subsets of CRC patients.
As previously reported, our cohort was significantly enriched with EOCRC, defined as < 50 years old [6]. We harnessed a robust, simple and affordable test called N_LyST to screen for probable Lynch, which fit into resource-limited settings in Indonesia [29]. The test is a polymerase chain reaction-high resolution melting analysis (PCR-HRMA)-based method, consisting of five mononucleotide markers for MSI, MLH1 promoter methylation and BRAF V600E mutation in a single PCR run. We found that there was a high proportion of probable Lynch in our Indonesian CRC cohort (13.85%) and it was strongly associated with EOCRC, despite still substantial numbers of EOCRC that were considered as non-LS/sporadic cases [6].
Similar to other studies, this study found no significant association between oncogenic KRAS and PIK3CA mutations and EOCRC [84]. Nonetheless, in this study, we found high KRAS and PIK3CA mutations, 81.25% and 59.38% respectively, in patients with probable Lynch status. These are much higher than previously reported in the literature, showing that KRAS mutations in the LS population range between 27% and 40% [27, 85-88]. We have reported the positive association of PIK3CA mutation with KRAS mutation [89]. There was a statistically significant association between probable Lynch status and KRAS mutation, but not PIK3CA mutation. It has been reported in the literature that KRAS mutation was more frequently found in LS-related MSI CRC as compared to sporadic MSI CRC [27, 86, 90, 91]. On the other hand, PIK3CA mutation was reported to be more common in somatically mutated MMR-deficient CRC [92].
We observed no differences in overall patients’ survival over a follow-up period of 120 months, between mutant and wild-type subgroups based on probable Lynch and non-LS/sporadic status for both KRAS and PIK3CA. Prior studies have shown that LS patients with CRC have a better prognosis than those with sporadic CRC, arguably due to its association with MSI and neoantigen generation in enhancing the immune response [93]. Although, as reported previously, we did not see this benefit of survival in our cohort [6]. It is appealing to speculate that high occurrences of KRAS mutation in our probable Lynch patients outweigh the survival benefit of MSI. To our knowledge, there have been no studies yet that examine the association of survival rates in KRAS and PIK3CA mutations with LS in CRC.
LS is highly heterogeneous, showing high variability in age at onset (despite enriched in EOCRC), penetrance of cancer, and clinical presentations, which may be partly attributable to the molecular profiles of carcinomas. As reviewed by Helderman et al (2021), LS heterogeneity is attributable to a variety of different molecular pathways of tumor development, and only partly depends on which MMR gene is mutated [94]. It is now recognized that LS CRCs develop via one of three pathways. The first pathway is adenoma-carcinoma pathway, in which adenomas develop independently of MMR deficiency. The second and third pathway is MMR-deficient crypt foci (MMR-DCF)-adenoma-carcinoma pathway and MMR-DCF-carcinoma pathway, which both start with MMR deficiency and is either followed by adenoma formation or results directly in a carcinoma [94, 95]. As widely known, APC mutations are more closely related to the development of adenomas, while CTNNB1 mutations appear to be associated with the MMR-DCF-carcinoma pathway [90, 94].
Recent studies have provided substantial evidence linking the methylation of MMR genes to the onset of LS [96, 97]. A significant portion of this evidence is derived from studies targeting the MLH1 gene, where methylation was observed in germline tissues of HNPCC patients who did not carry a germline mutation in the MLH1 gene [98]. Additionally, heritable germline epimutations in the MSH2 gene have been documented in LS families lacking MMR germline mutations [99]. A novel mechanism for inactivating the MSH2 gene has also been proposed. In several patients suspected of having LS but lacking detectable germline mutations in the MMR genes, researchers identified a heterozygous germline deletion encompassing the polyadenylation site within the final two exons of the epithelial cell adhesion molecule (EPCAM) gene [100]. Such deletion results disrupt the 3' end of the EPCAM gene, which induces transcriptional read-through. This aberrant transcription subsequently leads to epigenetic inactivation and silencing mechanisms that ultimately inhibit the proper expression of the MSH2 gene [101]. However, we acknowledge that we did not explore this mechanism in our study due to limited resources.
Understanding the molecular landscape of LS, such as RAF/MEK/ERK and PI3K/PTEN/AKT signaling, will allow more detailed stratification of LS patients and will facilitate the provision of optimal care to each patient, including the diagnosis, surveillance, and treatment. For instance, activating PIK3CA variants are potentially susceptible to preventive aspirin therapy by inducing the transcription of COX2 gene increasing the production of PGE2 [90, 102]. This is in addition to known resistance of anti-EGFR of CRC harboring KRAS, BRAF and PIK3CA mutations [103, 104]. However, it is not yet known whether cancers that develop during aspirin therapy have a specific molecular signature [105].
In summary, despite the limitations of this study, including being conducted in a single-center, tertiary hospital, while utilizing a low throughput (PCR-based) workflow for mutation detections, this study has contributed to the better understanding of CRC molecular features in an underrepresented population in current global literatures. The use of consecutive sampling at a tertiary hospital introduces potential ascertainment bias, likely over-representing those with advanced or treatment-resistant disease, as the center primarily receives referrals for patients who already underwent resection elsewhere. Acknowledging these limitations, future studies should include more representative samples from diverse settings and use alternative sampling methods to minimize bias. Our initial findings described in this paper and previous reports from our and other studies have indicated distinct genetic make-up such as high probability of LS, high frequency KRAS, and PIK3CA mutations, and lower BRAF mutation among CRC in Indonesia. This may underpin its unique clinical characteristics such as higher number of young patients and advanced disease stage. Further comprehensive multicenter analysis using high throughput techniques such as NGS is important to provide a more complete picture of the CRC carcinogenesis in Indonesia, with particular emphasis on EOCRC and LS. Understanding the global molecular landscape of CRC may reveal new knowledge that could challenge the current dogma, hence improving the effort to provide better care for the disease.
Conclusion
The high probability of LS in Indonesian CRC patients is associated with KRAS and PIK3CA mutations. It improved our understanding of CRC molecular features in an underrepresented population in global literature.
| Supplementary Material | ▴Top |
Suppl 1. Kaplan-Meier overall survival curves of CRC patients stratified by microsatellite instability (MSI) status. (A) Overall survival curves stratified by PIK3CA mutation and MSI status; (B) overall survival curves stratified by KRAS mutation and MSI status.
Suppl 2. Kaplan-Meier overall survival curves of CRC patients based on PIK3CA and KRAS mutation status. (A) Overall survival curves of PIK3CA mutant versus wild-type; (B) overall survival curves of KRAS mutant versus wild-type.
Suppl 3. Kaplan-Meier overall survival curve of CRC patients with Probable Lynch Syndrome versus sporadic cases.
Acknowledgments
The authors thank Aru W. Sudoyo and Ahmad Utomo for critical input into study design and analysis of results, Yasjudan Rastrama Putra and Yana Suryani for technical assistance and coordination.
Financial Disclosure
The authors received financial support from the Applied Research Grant 2020-2021, The Indonesian Ministry of Research, Technology, and Higher Education and the Institutional Links grant, ID 527558574, under the Newton Institutional Link-Indonesia KLN Fund partnership, funded by the UK Department for Business, Energy and Industrial Strategy and Indonesia Ministry of Research Technology & Higher Education and delivered by the British Council.
Conflict of Interest
MI was appointed as specialist committee member to the Diagnostics Assessment Committee of the National Institute for Health and Care Excellence (NICE), which produced the guidance DG27 on Lynch Syndrome testing. DSH, AMR, and MI are unpaid scientific advisors of PathGen Diagnostik Teknologi. Other authors have no competing interest to declare.
Informed Consent
All subjects provided written informed consent.
Author Contributions
Conception or design of the work: DSH, SHH, and SS; methodology: DSH, NY, GA, AMR, and SHH; software: GA, HH, MZA, AMR, WK, and SS; validation: DSH, NY, GA, AMR, SHH, and SS; formal analysis: DSH, NY, GA, HH, MZA, AMR, SHH, and SS; investigation: NY, GA, HH, AMR, WK, and SS; resources: DSH, NY, AYH, JK, SHH, and SS; data curation: DSH, NY, GA, VL, HH, ZH, ANG, AMR, and SS; writing-original draft preparation: DSH, VL, HH, ZH, ANG, and SS; writing-review and editing: DSH, AYH, MI, JK, SHH, and SS; visualization: GA, MZA, and SS; supervision: DSH, NY, MI, SHH, and SS; project administration: GA, VL, HH, ZH, and AMR; funding acquisition: DSH, MI, SHH, and SS; approval of the version to be published: all authors.
Data Availability
All data and related metadata underlying the findings reported in this manuscript have been provided as part of the submitted article. Any additional data that might support the findings of this study are available from the corresponding author, S.S., upon reasonable request.
| References | ▴Top |
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249.
doi pubmed - Khairina D, Suzanna E, Triana D, Kadir A, Widyastuti TH, Sulistyowati LS, et al. Profile of colorectal cancer in 14 provinces in Indonesia. J Glob Oncol. 2018;4(Supplement 2):66s.
- Abdullah M, Meilany S, Trimarsanto H, Malik SG, Sukartini N, Idrus F, Nursyirwan SA, et al. Genomic profiles of Indonesian colorectal cancer patients. F1000Res. 2022;11:443.
doi pubmed pmc - Idos G, Valle L. Lynch Syndrome. In: GeneReviews® [Internet]. University of Washington, Seattle; 2021.
- Bhattacharya P, McHugh TW. Lynch Syndrome. In: StatPearls [Internet]. StatPearls Publishing; 2023.
- Susanti S, Wibowo S, Akbariani G, Yoshuantari N, Heriyanto DS, Ridwanuloh AM, Hariyatun H, et al. Molecular analysis of colorectal cancers suggests a high frequency of lynch syndrome in Indonesia. Cancers (Basel). 2021;13(24):6245.
doi pubmed pmc - Singh MP, Rai S, Pandey A, Singh NK, Srivastava S. Molecular subtypes of colorectal cancer: An emerging therapeutic opportunity for personalized medicine. Genes Dis. 2021;8(2):133-145.
doi pubmed pmc - Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113-130.
doi pubmed - Malki A, ElRuz RA, Gupta I, Allouch A, Vranic S, Al Moustafa AE. Molecular mechanisms of colon cancer progression and metastasis: recent insights and advancements. Int J Mol Sci. 2020;22(1):130.
doi pubmed pmc - Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19-27.
pubmed pmc - Nyoman ADN, Suksmarini NMP, Pranata AAN, Rompis AY, Wayan Juli Sumadi I. The prevalence of KRAS and BRAF mutation in colorectal cancer patients in Bali. Indones J Biotechnol. 2022;27(1):29-35.
- Al-Sohaily S, Biankin A, Leong R, Kohonen-Corish M, Warusavitarne J. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol. 2012;27(9):1423-1431.
doi pubmed - Chen W, Frankel WL. A practical guide to biomarkers for the evaluation of colorectal cancer. Mod Pathol. 2019;32(Suppl 1):1-15.
doi pubmed - Carnevali IW, Cini G, Libera L, Sahnane N, Facchi S, Viel A, et al. Promoter methylation could be the second hit in lynch syndrome carcinogenesis. Genes [Internet]. 2023;14(11):2060.
doi - Peltomaki P, Nystrom M, Mecklin JP, Seppala TT. Lynch syndrome genetics and clinical implications. Gastroenterology. 2023;164(5):783-799.
doi pubmed - Newton K, Jorgensen NM, Wallace AJ, Buchanan DD, Lalloo F, McMahon RF, Hill J, et al. Tumour MLH1 promoter region methylation testing is an effective prescreen for Lynch Syndrome (HNPCC). J Med Genet. 2014;51(12):789-796.
doi pubmed pmc - Zhu G, Pei L, Xia H, Tang Q, Bi F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol Cancer. 2021;20(1):143.
doi pubmed pmc - Yang G, Yu XR, Weisenberger DJ, Lu T, Liang G. A multi-omics overview of colorectal cancer to address mechanisms of disease, metastasis, patient disparities and outcomes. Cancers (Basel). 2023;15(11):2934.
doi pubmed pmc - Meng M, Zhong K, Jiang T, Liu Z, Kwan HY, Su T. The current understanding on the impact of KRAS on colorectal cancer. Biomed Pharmacother. 2021;140:111717.
doi pubmed - Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, Cabiddu M, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015;51(5):587-594.
doi pubmed - Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH, Ghaedi K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019;9:97.
doi pubmed pmc - Lu X, Li Y, Li Y, Zhang X, Shi J, Feng H, Yu Z, et al. Prognostic and predictive biomarkers for anti-EGFR monoclonal antibody therapy in RAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. BMC Cancer. 2023;23(1):1117.
doi pubmed pmc - Luo Q, Chen D, Fan X, Fu X, Ma T, Chen D. KRAS and PIK3CA bi-mutations predict a poor prognosis in colorectal cancer patients: A single-site report. Transl Oncol. 2020;13(12):100874.
doi pubmed pmc - Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20(2):74-88.
doi pubmed pmc - Mirzapour Abbas Abadi Z, Samiee Rad F, Hamedi Asl D, Rahmani B, Soleimani Dodaran M, Peimani A. Clinicopathological Significance of PTEN Expression and Its Prognostic Effect in Colorectal Adenocarcinoma Patients. Iran J Pathol. 2022;17(2):150-158.
doi pubmed pmc - Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, Capelinha AF, et al. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26(1):158-163.
doi pubmed - Oliveira C, Westra JL, Arango D, Ollikainen M, Domingo E, Ferreira A, Velho S, et al. Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet. 2004;13(19):2303-2311.
doi pubmed - Ahn AR, Kim KM, Jang KY, Moon WS, Ha GW, Lee MR, Chung MJ. Correlation of PIK3CA mutation with programmed death ligand-1 (PD-L1) expression and their clinicopathological significance in colorectal cancer. Ann Transl Med. 2021;9(18):1406.
doi pubmed pmc - Susanti S, Fadhil W, Ebili HO, Asiri A, Nestarenkaite A, Hadjimichael E, Ham-Karim HA, et al. N_LyST: a simple and rapid screening test for Lynch syndrome. J Clin Pathol. 2018;71(8):713-720.
doi pubmed - Utomo A, Reimondos A, McDonald P, Utomo I, Hull T. Who wears the hijab? Predictors of veiling in greater Jakarta. Rev Relig Res [Internet]. 2018 [cited May 21, 2024]; Available from: https://journals.sagepub.com/doi/10.1007/s13644-018-0345-6.
- Wijaya UI, Nisfulaila I. Analisis implementasi matriks dalam estimasi laju pertumbuhan populasi wanita. Jurnal Riset Mahasiswa Matematika. 2024;3(2):101-109.
- Hassandi I. Millennials investment decision on Indonesia Government Sukuk: an analysis using behavioural factors. Jurnal Bisnis dan Manajemen (JBM). 2024;20(1);40-49.
- Permatasari A. Factors that affect rice crops price estimation based on grain mill enterprise in Ploso Jombang. Econ manag sustain. 2020;5(2):96-102.
- BPS Provinsi D.I. Yogyakarta [Internet]. [cited May 21, 2024]. Available from: https://yogyakarta.bps.go.id/publication/2024/02/28/8bf08007fc346b9f836ca663/provinsi-daerah-istimewa-yogyakarta-dalam-angka-2024.html.
- Fathoni MIA, Gunardi, Adi-Kusumo F, Hutajulu SH, Purwanto I. Characteristics of breast cancer patients at dr. Sardjito Hospital for early anticipation of neutropenia: Cross-sectional study. Ann Med Surg (Lond). 2022;73:103189.
doi pubmed pmc - Jeon SA, Ha YJ, Kim JH, Kim JH, Kim SK, Kim YS, Kim SY, et al. Genomic and transcriptomic analysis of Korean colorectal cancer patients. Genes Genomics. 2022;44(8):967-979.
doi pubmed pmc - Alkader MS, Altaha RZ, Badwan SA, Halalmeh AI, Al-Khawaldeh MH, Atmeh MT, Jabali EH, et al. Impact of KRAS mutation on survival outcome of patients with metastatic colorectal cancer in Jordan. Cureus. 2023;15(1):e33736.
doi pubmed pmc - Fu X, Huang Y, Fan X, Deng Y, Liu H, Zou H, Wu P, et al. Demographic trends and KRAS/BRAF(V600E) mutations in colorectal cancer patients of South China: A single-site report. Int J Cancer. 2019;144(9):2109-2117.
doi pubmed - Loong HH, Du N, Cheng C, Lin H, Guo J, Lin G, Li M, et al. KRAS G12C mutations in Asia: a landscape analysis of 11,951 Chinese tumor samples. Transl Lung Cancer Res. 2020;9(5):1759-1769.
doi pubmed pmc - Wong HL, Cui W, Loft M, Lee M, Wong R, Shapiro JD, et al. Assessing real-world outcomes in metastatic colorectal cancer with KRASG12C mutation. J Clin Oncol. 2020;38(15_suppl):e16072.
- El Agy F, El Bardai S, El Otmani I, Benbrahim Z, Karim MH, Mazaz K, Benjelloun EB, et al. Mutation status and prognostic value of KRAS and NRAS mutations in Moroccan colon cancer patients: A first report. PLoS One. 2021;16(3):e0248522.
doi pubmed pmc - Radanova M, Mihaylova G, Stoyanov GS, Draganova V, Zlatarov A, Kolev N, Dimitrova E, et al. KRAS mutation status in Bulgarian patients with advanced and metastatic colorectal cancer. Int J Mol Sci. 2023;24(16):12753.
doi pubmed pmc - Sanchez-Ibarra HE, Jiang X, Gallegos-Gonzalez EY, Cavazos-Gonzalez AC, Chen Y, Morcos F, Barrera-Saldana HA. KRAS, NRAS, and BRAF mutation prevalence, clinicopathological association, and their application in a predictive model in Mexican patients with metastatic colorectal cancer: A retrospective cohort study. PLoS One. 2020;15(7):e0235490.
doi pubmed pmc - Turpin A, Genin M, Hebbar M, Occelli F, Lanier C, Vasseur F, Descarpentries C, et al. Spatial heterogeneity of KRAS mutations in colorectal cancers in northern France. Cancer Manag Res. 2019;11:8337-8344.
doi pubmed pmc - Araujo LH, Souza BM, Leite LR, Parma SAF, Lopes NP, Malta FSV, Freire MCM. Molecular profile of KRAS G12C-mutant colorectal and non-small-cell lung cancer. BMC Cancer. 2021;21(1):193.
doi pubmed pmc - Gvaldin DY, Kit OI, Omelchuk EP, Kaymakchi DO, Poluektov SI, Petrov DS, et al. Frequency of somatic mutations in the KRAS gene in patients of the South Russia diagnosed with colorectal cancer. J Clin Oncol. 2019;37:e15081-e15081.
- Maus MK, Grimminger PP, Mack PC, Astrow SH, Stephens C, Zeger G, Hsiang J, et al. KRAS mutations in non-small-cell lung cancer and colorectal cancer: implications for EGFR-targeted therapies. Lung Cancer. 2014;83(2):163-167.
doi pubmed - Alghamdi M, Alabdullatif N, Al-Rashoud A, Alotaibi J, Alhussaini N, Elsirawani S, Somily H, et al. KRAS mutations in colorectal cancer: relationship with clinicopathological characteristics and impact on clinical outcomes in Saudi Arabia. Cureus. 2022;14(3):e23656.
doi pubmed pmc - Yang Q, Huo S, Sui Y, Du Z, Zhao H, Liu Y, Li W, et al. Mutation status and immunohistochemical correlation of KRAS, NRAS, and BRAF in 260 Chinese colorectal and gastric cancers. Front Oncol. 2018;8:487.
doi pubmed pmc - Rahadiani N, Handjari DR, Stephanie M, Krisnuhoni E. The low prevalence of colonic serrated adenocarcinoma with high KRAS mutational status at Cipto Mangunkusumo Hospital, Indonesia. Med J Indones. 2018;27(3):161-168.
- Stefanius K, Ylitalo L, Tuomisto A, Kuivila R, Kantola T, Sirnio P, Karttunen TJ, et al. Frequent mutations of KRAS in addition to BRAF in colorectal serrated adenocarcinoma. Histopathology. 2011;58(5):679-692.
doi pubmed pmc - Garcia-Solano J, Conesa-Zamora P, Carbonell P, Trujillo-Santos J, Torres-Moreno DD, Pagan-Gomez I, Rodriguez-Braun E, et al. Colorectal serrated adenocarcinoma shows a different profile of oncogene mutations, MSI status and DNA repair protein expression compared to conventional and sporadic MSI-H carcinomas. Int J Cancer. 2012;131(8):1790-1799.
doi pubmed - Marbun VMG, Erlina L, Lalisang TJM. Genomic landscape of pathogenic mutation of APC, KRAS, TP53, PIK3CA, and MLH1 in Indonesian colorectal cancer. PLoS One. 2022;17(6):e0267090.
doi pubmed pmc - He K, Wang Y, Zhong Y, Pan X, Si L, Lu J. KRAS codon 12 mutation is associated with more aggressive invasiveness in synchronous metastatic colorectal cancer (mCRC): retrospective research. Onco Targets Ther. 2020;13:12601-12613.
doi pubmed pmc - Heuvelings DJI, Wintjens A, Luyten J, Wilmink G, Moonen L, Speel EM, de Hingh I, et al. DNA and RNA alterations associated with colorectal peritoneal metastases: a systematic review. Cancers (Basel). 2023;15(2):549.
doi pubmed pmc - Li Y, Xiao J, Zhang T, Zheng Y, Jin H. Analysis of KRAS, NRAS, and BRAF mutations, microsatellite instability, and relevant prognosis effects in patients with early colorectal cancer: a cohort study in East Asia. Front Oncol. 2022;12:897548.
doi pubmed pmc - Chang XN, Shang FM, Jiang HY, Chen C, Zhao ZY, Deng SH, Fan J, et al. Clinicopathological features and prognostic value of KRAS/NRAS/BRAF mutations in colorectal cancer patients of central China. Curr Med Sci. 2021;41(1):118-126.
doi pubmed - Mei WJ, Mi M, Qian J, Xiao N, Yuan Y, Ding PR. Clinicopathological characteristics of high microsatellite instability/mismatch repair-deficient colorectal cancer: A narrative review. Front Immunol. 2022;13:1019582.
doi pubmed pmc - Ye ZL, Qiu MZ, Tang T, Wang F, Zhou YX, Lei MJ, Guan WL, et al. Gene mutation profiling in Chinese colorectal cancer patients and its association with clinicopathological characteristics and prognosis. Cancer Med. 2020;9(2):745-756.
doi pubmed pmc - Ye JX, Liu Y, Qin Y, Zhong HH, Yi WN, Shi XY. KRAS and BRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World J Gastroenterol. 2015;21(5):1595-1605.
doi pubmed pmc - Molina-Cerrillo J, San Roman M, Pozas J, Alonso-Gordoa T, Pozas M, Conde E, Rosas M, et al. BRAF Mutated Colorectal Cancer: New Treatment Approaches. Cancers (Basel). 2020;12(6):1571.
doi pubmed pmc - Kassem NM, Emera G, Kassem HA, Medhat N, Nagdy B, Tareq M, et al. Clinicopathological features of Egyptian colorectal cancer patients regarding somatic genetic mutations especially in KRAS gene and microsatellite instability status: a pilot study. Egypt J Med Hum Genet [Internet]. 2019;20(1);20.
- Geiersbach KB, Samowitz WS. Microsatellite instability and colorectal cancer. Arch Pathol Lab Med. 2011;135(10):1269-1277.
doi pubmed - Uhlig J, Cecchini M, Sheth A, Stein S, Lacy J, Kim HS. Microsatellite Instability and KRAS Mutation in Stage IV Colorectal Cancer: Prevalence, Geographic Discrepancies, and Outcomes From the National Cancer Database. J Natl Compr Canc Netw. 2021;19(3):307-318.
doi pubmed - Zhao Y, Miyashita K, Ando T, Kakeji Y, Yamanaka T, Taguchi K, Ushijima T, et al. Exclusive KRAS mutation in microsatellite-unstable human colorectal carcinomas with sequence alterations in the DNA mismatch repair gene, MLH1. Gene. 2008;423(2):188-193.
doi pubmed - Cushman-Vokoun AM, Stover DG, Zhao Z, Koehler EA, Berlin JD, Vnencak-Jones CL. Clinical utility of KRAS and BRAF mutations in a cohort of patients with colorectal neoplasms submitted for microsatellite instability testing. Clin Colorectal Cancer. 2013;12(3):168-178.
doi pubmed pmc - Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105(15):1151-1156.
doi pubmed pmc - Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65(14):6063-6069.
doi pubmed - Asaka S, Arai Y, Nishimura Y, Yamaguchi K, Ishikubo T, Yatsuoka T, Tanaka Y, et al. Microsatellite instability-low colorectal cancer acquires a KRAS mutation during the progression from Dukes' A to Dukes' B. Carcinogenesis. 2009;30(3):494-499.
doi pubmed - Gong J, Cho M, Sy M, Salgia R, Fakih M. Molecular profiling of metastatic colorectal tumors using next-generation sequencing: a single-institution experience. Oncotarget. 2017;8(26):42198-42213.
doi pubmed pmc - Midthun L, Shaheen S, Deisch J, Senthil M, Tsai J, Hsueh CT. Concomitant KRAS and BRAF mutations in colorectal cancer. J Gastrointest Oncol. 2019;10(3):577-581.
doi pubmed pmc - Xie MZ, Li JL, Cai ZM, Li KZ, Hu BL. Impact of primary colorectal Cancer location on the KRAS status and its prognostic value. BMC Gastroenterol. 2019;19(1):46.
doi pubmed pmc - Bonnot PE, Passot G. RAS mutation: site of disease and recurrence pattern in colorectal cancer. Chin Clin Oncol. 2019;8(5):55.
doi pubmed - Pirvu EE, Severin E, Patru RI, Nita I, Toma SA, Macarie RR, Cocioaba CE, et al. Correlations between demographic, clinical, and paraclinical variables and outcomes in patients with KRAS-Mutant or KRAS wild-type metastatic colorectal cancer - a retrospective study from a tertiary-level center in Romania. Diagnostics (Basel). 2023;13(18):2930.
doi pubmed pmc - Oliveira-Silveira J, Filippi-Chiela E, Saffi J. Laterality influence on gene expression of DNA damage repair in colorectal cancer. Sci Rep. 2023;13(1):15963.
doi pubmed pmc - Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference between left-sided and right-sided colorectal cancer: a focused review of literature. Gastroenterology Res. 2018;11(4):264-273.
doi pubmed pmc - Labeda I, Syarifuddin E, Uwuratuw J, Pattelongi I, Faruk M. Analysis of pik3ca expression to clinicopathology features of colorectal cancer in Makassar, Indonesia. Int J Med Robot. 2020;6(1):48-52.
- Jin J, Shi Y, Zhang S, Yang S. PIK3CA mutation and clinicopathological features of colorectal cancer: a systematic review and Meta-Analysis. Acta Oncol. 2020;59(1):66-74.
doi pubmed - Westdorp H, Fennemann FL, Weren RD, Bisseling TM, Ligtenberg MJ, Figdor CG, Schreibelt G, et al. Opportunities for immunotherapy in microsatellite instable colorectal cancer. Cancer Immunol Immunother. 2016;65(10):1249-1259.
doi pubmed pmc - Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front Immunol. 2020;11:369.
doi pubmed pmc - Koury J, Lucero M, Cato C, Chang L, Geiger J, Henry D, Hernandez J, et al. Immunotherapies: exploiting the immune system for cancer treatment. J Immunol Res. 2018;2018:9585614.
doi pubmed pmc - Nusrat M, Syed MA, Katkhuda R, Parra ER, Wistuba II, Kong P, et al. The immune impact of PI3K-AKT pathway inhibition in colorectal cancer. J Clin Oncol [Internet]. 2022;40:154.
- Li W, Qiu T, Dong L, Zhang F, Guo L, Ying J. Prevalence and characteristics of PIK3CA mutation in mismatch repair-deficient colorectal cancer. J Cancer. 2020;11(13):3827-3833.
doi pubmed pmc - Hamilton AC, Bannon FJ, Dunne PD, James J, McQuaid S, Gray RT et al. Distinct molecular profiles of sporadic early-onset colorectal cancer: a population-based cohort and systematic review. Gastro Hep Advances. 2023;2(3):347-359.
- Garre P, Martin L, Bando I, Tosar A, Llovet P, Sanz J, Romero A, et al. Cancer risk and overall survival in mismatch repair proficient hereditary non-polyposis colorectal cancer, Lynch syndrome and sporadic colorectal cancer. Fam Cancer. 2014;13(1):109-119.
doi pubmed - Li W, Zhi W, Zou S, Qiu T, Ling Y, Shan L, Shi S, et al. Distinct clinicopathological patterns of mismatch repair status in colorectal cancer stratified by KRAS mutations. PLoS One. 2015;10(6):e0128202.
doi pubmed pmc - Abdel-Rahman WM, Ollikainen M, Kariola R, Jarvinen HJ, Mecklin JP, Nystrom-Lahti M, Knuutila S, et al. Comprehensive characterization of HNPCC-related colorectal cancers reveals striking molecular features in families with no germline mismatch repair gene mutations. Oncogene. 2005;24(9):1542-1551.
doi pubmed - Goel A, Xicola RM, Nguyen TP, Doyle BJ, Sohn VR, Bandipalliam P, Rozek LS, et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology. 2010;138(5):1854-1862.
doi pubmed pmc - Susanti S, Fadhil W, Murtaza S, Hassall JC, Ebili HO, Oniscu A, Ilyas M. Positive association of PIK3CA mutation with KRAS mutation but not BRAF mutation in colorectal cancer suggests co-selection is gene specific but not pathway specific. J Clin Pathol. 2019;72(3):263-264.
doi pubmed - Ahadova A, Gallon R, Gebert J, Ballhausen A, Endris V, Kirchner M, Stenzinger A, et al. Three molecular pathways model colorectal carcinogenesis in Lynch syndrome. Int J Cancer. 2018;143(1):139-150.
doi pubmed - Ten Broeke SW, van der Klift HM, Tops CMJ, Aretz S, Bernstein I, Buchanan DD, de la Chapelle A, et al. Cancer risks for PMS2-associated lynch syndrome. J Clin Oncol. 2018;36(29):2961-2968.
doi pubmed pmc - Cohen SA, Turner EH, Beightol MB, Jacobson A, Gooley TA, Salipante SJ, Haraldsdottir S, et al. Frequent PIK3CA mutations in colorectal and endometrial tumors with 2 or more somatic mutations in mismatch repair genes. Gastroenterology. 2016;151(3):440-447 e441.
doi pubmed pmc - Battaglin F, Naseem M, Lenz HJ, Salem ME. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol. 2018;16(11):735-745.
pubmed pmc - Helderman NC, Bajwa-Ten Broeke SW, Morreau H, Suerink M, Terlouw D, van der Werf TLAS, van Wezel T, et al. The diverse molecular profiles of lynch syndrome-associated colorectal cancers are (highly) dependent on underlying germline mismatch repair mutations. Crit Rev Oncol Hematol. 2021;163(103338.
doi pubmed - Lepore Signorile M, Disciglio V, Di Carlo G, Pisani A, Simone C, Ingravallo G. From Genetics to Histomolecular Characterization: An Insight into Colorectal Carcinogenesis in Lynch Syndrome. Int J Mol Sci. 2021;22(13):6767.
doi pubmed pmc - Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, Ward RL. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356(7):697-705.
doi pubmed - Nagasaka T, Rhees J, Kloor M, Gebert J, Naomoto Y, Boland CR, Goel A. Somatic hypermethylation of MSH2 is a frequent event in Lynch Syndrome colorectal cancers. Cancer Res. 2010;70(8):3098-3108.
doi pubmed pmc - Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62(14):3925-3928.
pubmed - Kovacs ME, Papp J, Szentirmay Z, Otto S, Olah E. Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat. 2009;30(2):197-203.
doi pubmed - Guarinos C, Castillejo A, Barbera VM, Perez-Carbonell L, Sanchez-Heras AB, Segura A, Guillen-Ponce C, et al. EPCAM germ line deletions as causes of Lynch syndrome in Spanish patients. J Mol Diagn. 2010;12(6):765-770.
doi pubmed pmc - Kempers MJ, Kuiper RP, Ockeloen CW, Chappuis PO, Hutter P, Rahner N, Schackert HK, et al. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: a cohort study. Lancet Oncol. 2011;12(1):49-55.
doi pubmed pmc - Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149(7):1884-1895.e1884.
doi pubmed pmc - Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, Yu Q, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2017;8(3):3980-4000.
doi pubmed pmc - Li ZN, Zhao L, Yu LF, Wei MJ. BRAF and KRAS mutations in metastatic colorectal cancer: future perspectives for personalized therapy. Gastroenterol Rep (Oxf). 2020;8(3):192-205.
doi pubmed pmc - Burn J, Sheth H, Elliott F, Reed L, Macrae F, Mecklin JP, Moslein G, et al. Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: a double-blind, randomised, placebo-controlled trial. Lancet. 2020;395(10240):1855-1863.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


