| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 5, October 2024, pages 801-808
Patterns and Frequency of Pathogenic Germline Variants Among Prostate Cancer Patients Utilizing Multi-Gene Panel Genetic Testing
Ramiz Abu Hijliha, Baha Sharafb, Samer Salahb, Hira Bani Hanib, Sarah M. Nielsenc, Brandie Healdc, Edward D. Esplinc, Rami Ghanemd, Abdulla Alzibdeha, Tamer Al-Batshb, Yosra Al-Masrib, Hikmat Abdel-Razeqb, e, f
aDepartment of Radiation Oncology, King Hussein Cancer Center, Amman, Jordan
bDepartment of Internal Medicine, King Hussein Cancer Center, Amman, Jordan
cInvitae Corporation, San Francisco, CA, USA
dDepartment of Surgery, King Hussein Cancer Center, Amman, Jordan
eSchool of Medicine, the University of Jordan, Amman, Jordan
fCorresponding Author: Hikmat Abdel-Razeq, Department of Internal Medicine, King Hussein Cancer Center, Amman 11941, Jordan
Manuscript submitted May 19, 2024, accepted June 21, 2024, published online July 18, 2024
Short title: Germline Genetic Testing for Prostate Cancer
doi: https://doi.org/10.14740/wjon1896
| Abstract | ▴Top |
Background: Germline genetic testing (GGT) has significant implications in the management of patients with prostate cancer (PCa). Herein, we report on patterns and frequency of pathogenic/likely pathogenic germline variants (P/LPGVs) among newly diagnosed Arab patients with PCa.
Methods: Patients meeting the National Comprehensive Cancer Network (NCCN) eligibility criteria for GGT were offered a 19-gene PCa panel or an expanded 84-gene multi-cancer panel.
Results: During the study period, 231 patients were enrolled; 107 (46.3%) had metastatic disease at diagnosis. In total, 17 P/LPGVs were detected in 17 patients (7.4%). Among the 113 (48.9%) patients who underwent GGT with the 19-gene panel, eight (7.1%) had P/LPGVs, compared to nine (7.6%) of the 118 (51.1%) who did GGT through the expanded 84-gene panel (P = 0.88). Variant of uncertain significance (VUS) rate was higher (n = 73, 61.9%) among the group who underwent expanded 84-gene panel testing compared to those who underwent the 19-gene PCa panel (n = 35, 30.9%) (P = 0.001). P/LPGVs in DNA damage repair (DDR) genes, most frequently BRCA2, CHEK2 and TP53, were the most common P/LPGVs findings.
Conclusion: This study is the first to characterize the germline genetic profile of an Arab population with PCa. All detected P/LPGVs were potentially actionable, with most variants able to be detected with a PCa-specific panel.
Keywords: Prostate cancer; Germline testing; Pathogenic mutations; VUS; BRCA
| Introduction | ▴Top |
Prostate cancer (PCa) is the second most common cancer diagnosed among men worldwide and its incidence is highest in Western societies [1]. Treatment options for localized disease include active surveillance, radical prostatectomy, or radiotherapy. Selection of the treatment modality is typically dependent on a multi-disciplinary approach that incorporates risk category, performance status, associated comorbidities, and patients’ preference [1, 2]. The standard of care for patients with metastatic disease has shifted towards early therapy intensification, which involves combining androgen deprivation therapy (ADT) with either docetaxel or a novel oral androgen signaling inhibitor. This approach has demonstrated improved outcomes compared to ADT alone [2-5].
Despite the progress made in treating localized and metastatic PCa, a considerable number of patients still experience poor outcomes. Expanding our knowledge of molecular biomarkers and pathogenic/likely pathogenic germline variants (P/LPGVs) holds promise in revealing valuable insights into the diverse characteristics and outcomes of these patients, and in informing treatment decisions. New studies are shedding light on multiple genomic and environmental risk factors in PCa. Nevertheless, current understanding of the influence of these factors on outcomes is still limited [6].
Over the last decade, there has been a heightened interest in the inherited component of PCa, as these P/LPGVs may have implications on screening and treatment decisions [7-9]. Germline genetic testing (GGT) identifies a spectrum of cancer predisposition variants, from low to high risk. At least 170 inherited variants have been identified, which are involved in about one-third of familial PCa cases. This list includes DNA damage repair (DDR) genes such as ATM, BRCA1, BRCA2, CHEK2, MLH1, MSH2, MSH6, NBN, PALB2 and PMS2 [8-10], some of which are predictive of unfavorable prognosis irrespective of disease stage and may guide treatment selection, such as the use of poly-ADP ribose polymerase (PARP) inhibitors in patients with metastatic PCa who harbor germline BRCA1/2 mutations [8-12]. Accordingly, current guidelines, including the National Comprehensive Cancer Network (NCCN), the Philadelphia expert consensus (2019) and the American Urological Association (AUA), endorse GGT for PCa with minor differences between these guidelines regarding the panel of genes to be tested and the eligibility criteria [13-15]. Currently, NCCN guidelines recommend GGT for patients with high-risk localized, locally advanced and metastatic disease, or for patients with positive family history for cancer such as prostate and hereditary breast and ovarian syndromes, regardless of the disease stage [13].
The distribution of many P/LPGVs varies according to ethnicity and geographic region; however, data on the P/LPGV landscape of PCa patients from the Middle East region remain scarce. Herein, we prospectively investigated patients who were treated at King Hussein Cancer Center, aiming to identify the patterns and frequency of P/LPGVs in this population. Moreover, we sought to explore the additional value of expanded multi-gene panel testing (MGPT) for detection of additional P/LPGVs not captured with more limited MGPT.
| Materials and Methods | ▴Top |
Patient population
This was a prospective, single-institution study of P/LPGVs among newly diagnosed patients with prostatic adenocarcinoma treated at King Hussein Cancer Center from March 2021 to July 2022. Eligibility criteria included: age ≥ 18 years of Arab descent who met the NCCN criteria (version 1.2020) for GGT. Eligible men were stratified according to castrate status as per NCCN definition; castration resistance is defined as any biochemical, radiographic, or clinical progression, with serum testosterone < 50 ng/mL after hormonal therapy [16].
Genetic counseling
Patients were approached through the genetic counseling clinic, and those who met the NCCN criteria were offered GGT with either a 19-gene PCa panel (“prostate MGPT”) or an expanded 84-gene multi-cancer panel (“multi-cancer MGPT”), per patient choice. The prostate MGPT consists of 10 genes with established evidence for PCa and recommended by NCCN (ATM, BRCA1, BRCA2, CHEK2, HOXB13, MLH1, MSH2, MSH6, PALB2, and PMS2), in addition to nine other genes with preliminary evidence for PCa risk (EPCAM, NBN, TP53, ATR, BRIP1, FANCA, GEN1, RAD51C and RAD51D). The multi-cancer MGPT consists of 84 genes associated with various hereditary cancer syndromes (Supplementary Material 1, www.wjon.org). Full-gene sequencing, deletion/duplication analysis, and variant interpretation were performed at a single commercial laboratory (Invitae Corporation, San Francisco, CA, USA), as previously described [17, 18]. Results were reported according to the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) five tiers classification: pathogenic (P), likely pathogenic (LP), variant of uncertain significance (VUS), likely benign and benign. Variants were classified as positive (≥ 1 P/LP), VUS (≥ 1 VUS in the absence of a P/LPGV) and negative (no P/LP or VUS identified). Carriers of single P/LPGV in genes associated with autosomal recessive inheritance (e.g., MUTYH) were not counted in the overall P/LPGV yield. NCCN guidelines recommend GGT for patients with high-risk localized and metastatic disease, or for patients with positive family history for cancer such as prostate and hereditary breast and ovarian syndromes, regardless of the disease stage [13]. Results were sent to the treating physician and the genetic counselor disclosed results to the patient.
Genetic testing
Patients’ identifiers were coded, and all GGT was performed on DNA extracted using a peripheral blood sample at a reference laboratory (Invitae Corporation) for both the limited and the expanded 84-gene MGP. Whole gene sequencing was performed utilizing a next-generation sequencing (NGS) platform as previously described [17, 18].
Data collection and study procedures
Electronic medical records were accessed to obtain demographics and clinical data. Cancer family history was obtained during the genetic counseling consultation. Demographics, clinical characteristics, cancer family history and GGT results were documented in a Health Insurance Portability and Accountability Act (HIPAA)-compliant database. Research was performed in accordance with relevant local and international guidelines and regulations including the Declaration of Helsinki. The study was approved by the King Hussein Cancer Center Institutional Review Board (IRB) and registered under Clinicaltrial.gov identifier: NCT04920513.
Statistical analysis
Descriptive statistics were utilized to report means, median, standard deviations, and proportions. The Chi-square test was used to compare the rate of detection of P/LPGV and VUS and to explore the correlation between disease characteristics and the likelihood of identifying a P/LPGV; all P- values less than 0.05 were considered statistically significant. All statistical analyses were performed using SPSS version 19 (SPSS Inc., Chicago, IL).
| Results | ▴Top |
Patient characteristics
A total of 266 patients were identified as eligible for GGT. After providing genetic counseling, 35 (13.1%) patients declined GGT; the majority (n = 24; 68.6%) did not believe in the value of GGT while seven (20.0%) were concerned about the stress related to results and an additional four (11.4%) declined testing for financial reasons. The remaining 231 patients underwent GGT and comprised the cohort for analysis. The median age at diagnosis was 67 (range, 45 - 86) years, 124 (53.7%) patients had localized disease while 107 (46.3%) had metastatic disease. The median baseline serum prostate-specific antigen (PSA) value was 32 (range, 2.4 - 3,975) ng/mL, and 146 (63.2%) of patients had a Gleason score ≥ 8. The most frequent indication for GGT was localized high-risk disease in 115 (49.8%) or metastatic in 107 (46.3%) and nine (3.9%) with positive family history for cancer. A total of 113 (48.9%) patients elected to do prostate MGPT, while the other 118 (51.1%) patients opted for multi-cancer MGPT. Both groups were balanced in terms of their clinical characteristics.
Genetic testing results
A total of 17 P/LPGV were identified in 17 (7.4%) patients. Among the 113 patients who underwent prostate MGPT, eight (7.1%) had P/LPGV, compared to nine (7.6%) among the 118 who had multi-cancer MGPT (P = 0.88). P/LPGVs were as follows: 14 patients (82.4%) had P/LPGV in DDR genes including ATM (n = 1), BRCA2 (n = 5), BRIP1 (n = 1), CHEK2 (n = 2), MLH1 (n = 1), PALB2 (n=1), PMS2 (n = 1), and TP53 (n = 2). Additional P/LPGV findings included RET (n = 1) and APC I1307K (n = 2) (Fig. 1). Notably, an additional five patients, not included in the total P/LPGV yield above, were identified as carriers of a single P/LPGV in a gene associated with an autosomal recessive cancer syndrome (one patient each with MSH3, NBN, RAD50, RECQL4, and WRN). Of these alterations, WRN (Intron 18; c.2088+1G>T (splice donor)) and MSH3 (deletion exon 9-19) have not been previously documented in ClinVar (Supplementary Material 2, www.wjon.org).
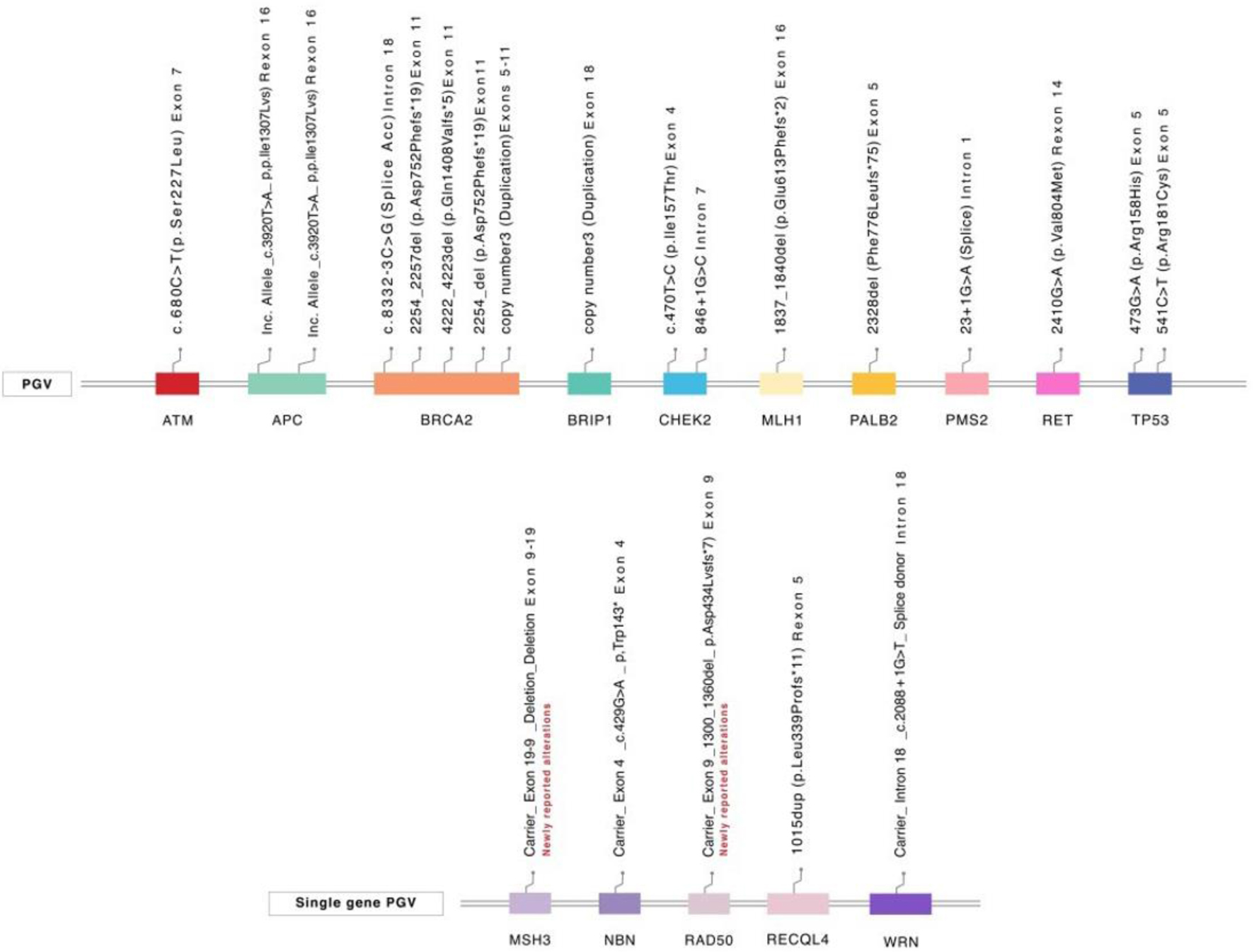 Click for large image | Figure 1. Pathogenic/likely pathogenic variants. PGV: pathogenic germline variant. |
Only three of the detected P/LPGVs were not included in the PCa MGPT (RET and APC). Notably, there were no significant differences in P/LPGV frequency by various clinical characteristics, including castration status (sensitive vs. resistant), age (≤ 65 vs. > 65), Gleason score (≥ 8 vs. < 8), disease stage (localized vs. metastatic) or family history (Table 1). All detected P/LPGVs were actionable based on published management guidelines (n = 17) or potential eligibility for clinical trials and/or targeted therapies (n = 12).
 Click to view | Table 1. PGV Rates Across Clinical Variables (n = 231) |
VUS
The most frequently observed alterations in the whole cohort were VUS in 108 (46.8%) patients with the absence of a P/PLGV. VUSs were more commonly detected with the multi-cancer MGPT (n = 73, 61.9%), compared to the PCa MGPT (n = 35, 30.9%) (P < 0.001).
| Discussion | ▴Top |
To the best of our knowledge, this is the first study to examine the prevalence and characteristics of P/LPGV in Arab PCa patients. The first comprehensive GGT study in PCa was published by Pritchard et al in 2016. Their study examined 692 patients with metastatic disease, who were unselected by family history or age at diagnosis. The study reported an 11.8% prevalence of P/LPGV in DNA repair genes. Notably, this frequency was significantly higher than that observed in a cohort of 499 patients with localized disease, where only 4.9% had P/LPGV in DNA repair genes [19]. Subsequent studies, including both retrospective and prospective unselected cohorts, have reported P/LPGV prevalence ranging from 8% to 20% [20, 21].
The prevalence of P/LPGV in our patient population is slightly lower (7.4%) than has been reported in the aforementioned studies [22-24]; however, it is similar to the yield recently reported by Shore et al (7.7%) in a prospective GGT study of unselected PCa where the majority had low-grade localized disease [25]. Similar to our study, their P/LPGV yield excluded carriers of single P/LPGV in autosomal recessive cancer syndrome genes. Various factors contribute to the range of P/LPGV identified, including study design whether retrospective or prospective, patient characteristics, including the proportion of metastatic or advanced disease, in addition to specific criteria used for GGT eligibility, and differences in testing methodologies or genes included on MGPT. Furthermore, racial disparities in P/LPGV rates may contribute to this variation. A recent study by Giri et al highlighted the discrepancy in P/LPGV rates between PCa patients of different ethnicities in the USA. The frequency of P/LPGV was notably higher in White patients (11%) compared to Black/African American patients (5.9%) [21]. This is not unexpected given the over-representation of White European participants in the genetic studies that inform GGT criteria and variant interpretation databases, resulting in higher rates of uncertain findings and lower rates of definitive molecular diagnoses in individuals from historically underrepresented non-White populations, including Arab patients [26-28]. This is evident in a study from Korea which reported P/LPGV in 5.8% of tested Korean men with mutations similar to our study [29]. Furthermore, many studies have documented higher P/LPGV prevalence in metastatic castrate-resistant PCa (mCRPC) compared with castrate-sensitive cases, nevertheless, our results did not show a significant difference between the two groups, though this may be due to our study being under-powered to detect the difference between these two groups.
Consistent with other data from literature [20, 21, 30], our study showed that the most prevalent P/LPGVs in PCa were in homologous recombination repair (HRR) genes, whereas only 0.9% had DNA mismatch repair (MMR) gene alterations. Notably, we reported two cases (12% of positive results) of the increased risk allele APC I1307K, which is associated with increased risk for colorectal cancer and is prevalent among Ashkenazi Jews [24]; the high incidence in the Arab population warrants further investigation. We acknowledge that it is controversial to classify mutations such as APC I1307K CHEK2 c.470T>C as pathogenic, given their high prevalence in general population. This is another hot area for future research.
The NCCN PCa GGT guidelines (version 4.2023) recommend testing of 10 genes; however, most commercial laboratories offer customized panels with specific add-on genes with preliminary evidence. The prostate MGPT we utilized for testing included nine additional non-NCCN genes, which resulted in the identification of three additional P/LPGVs. Multi-cancer MPGT resulted in the detection of only three additional P/LPGVs (two APC and one RET with RECQL4 as a carrier finding). Multi-cancer MGPT was associated with a statistically significant increase in VUS. However, patients with VUS should be managed based on their personal and family history as the majority of VUSs are ultimately reclassified to benign [31-33]. Nevertheless, detecting these VUSs in understudied population, such as our population, might result in reclassification of some of these genes to P/LPGV. Meanwhile, VUS can significantly increase anxiety and psychological distress for both patients and physicians, because of the uncertainty of VUS impacts. However, knowledge accumulation would be useful in future genetic research, moreover patients and families with VUS can be notified when the gene is reclassified.
Notably, four patients were found to have alteration in a gene associated with autosomal recessive cancer inheritance. While these findings may not directly impact cancer risk, they can be important for cascade testing, and in the case of DDR genes such as NBN and RAD50, may even qualify patients for clinical trials (NCT03413995, NCT03209401). Interestingly, the WRN (splice site) and MSH3 (deletion) variants detected in our study have not been reported in the literature or public databases. Furthermore, the other two identified P/LPGVs (RAD50 and NBN) have been reported in breast cancer but not in PCa [22, 23]. Additional studies are warranted to assess the significance of these alterations in PCa. Another observation in our study was the TP53 mutation in the context of Li-Fraumeni spectrum cancers, as the two patients with TP53 had family history for malignancy. One had family history of lung, breast, colorectal, lymphoma and kidney cancers in first-degree relatives. While the second had family history of breast cancer and PCa in three of his relatives.
GGT plays a crucial role in precision medicine, by providing relevant prognostic information and identifying gene alterations that could serve as therapeutic targets. Accumulating data confirm the clinical activity of PARP inhibitors in patients with germline and somatic DDR gene alterations [8, 34, 35]. In the PROfound trial, olaparib showed improved radiographic progression-free survival (rPFS) and overall survival (OS) in mCRPC compared to novel hormonal agents, particularly in patients with BRCA1/2 or ATM alterations, and these findings led to FDA approval in May 2020 [36, 37]. Another trial, TRITON 3, evaluated rucaparib in mCRPC patients with deleterious alterations in BRCA1, BRCA2, or ATM, showing improved rPFS compared to investigator’s choice treatment in patients with BRCA1/BRCA2 alterations but not in those with ATM alterations [35]. Recently, the FDA approved the combination of olaparib and abiraterone with prednisolone in patients with BRCA-mutated mCRPC based on data from the PROPEL trial, demonstrating improved rPFS and OS compared to abiraterone and prednisolone. This improvement was observed in patients with BRCA mutations but not in those without BRCA mutations [38]. More recently, niraparib has been approved in combination with abiraterone in first line setting for somatic BRCA-mutated mCRPC based on the phase 3 MAGNITUDE study [39-41]. Noteworthy, these trials have mainly included somatic rather than germline mutations.
The prevalence of MMR P/LPGV in advanced PCa has been reported to be 0.5-1.7% [8, 42-45]. Thus, our 0.9% rate of MMR genes is consistent with the literature. However, the two patients with MMR aberrations in our cohort had alterations in MLH1 and PMS2 genes, in contrast to the MSH2 and MSH6 P/LPGV most commonly reported in other PCa studies [42-44]. Current evidence shows a two to threefold higher frequency of MMR mutations in metastatic PCa compared to localized disease [45]. Aside from the advanced disease stage, there is a higher likelihood of identifying MMR P/LPGV in patients with Gleason score ≥ 8 and ductal histologic variants [46, 47]. Up to 12% of patients with metastatic PCa are found to have somatic MMR gene mutations and/or microsatellite instability (MSI)-high status [45]. Identification of a P/LPGV in an MMR gene is often an indication that the tumor may be MMR-deficient, and the patient may therefore be eligible for treatment with the FDA-approved immune checkpoint inhibitor pembrolizumab [48, 49].
In our study, we found that all identified P/LPGVs, most of which were in the DDR pathway, were actionable based on published management guidelines, or potential eligibility for clinical trials and/or targeted therapies. This suggests that patients carrying these genes may potentially benefit from novel drugs. These variants may also play a crucial role in determining disease prognosis and personalized management plans. Moreover, the detection of these P/LPGVs enables cancer screening and prevention of the patients and their families.
We conducted prospective GGT of 231 men. However, it is worth noting that the majority of men with PCa in our region are not offered GGT, even when they meet the recommendations set by major professional societies. This disparity can be attributed to many factors such as a lack of genetic counselors, insufficient knowledge among treating physicians, and a lack of insurance coverage. It is important to acknowledge that these barriers are not limited to our region alone. A survey conducted by the Prostate Cancer Clinical Trial Consortium revealed that less than 40% of treating physicians in academic centers refer eligible candidates for GGT, and only 12% of individuals with high-risk localized disease were considered for testing [50]. In our study, we encountered a refusal rate of 13% among patients, and interestingly, the main reasons for this were disbelief in the value of GGT, and concern about the social and psychological impact of receiving positive results. These findings warrant further investigation and targeted interventions.
Finally, we acknowledge some limitations relevant to our study. First, our study did not include disease and treatment outcomes, which could have provided valuable insights into the correlation between GGT and patients’ prognosis. Second, although our cohort included a significant number of patients relative to disease prevalence in our region, it is still a small cohort compared to other studies in the literature. Third, we failed to identify a significant correlation between many patient and disease characteristics and the likelihood of increased detection of P/LPGV, which could require a larger sample size. Nevertheless, this study contributes valuable information to the literature regarding the germline genetic profile of Arab patients with PCa.
Conclusions
Our study represents the first report of GGT conducted in an Arab population with PCa. Among patients tested, 7.4% exhibited P/LPGV, mostly involving DDR genes. We observed that a panel of 19 prostate-specific genes was effective in identifying the majority of P/LPGV. More studies are needed to correlate these GGT results with clinical outcomes in our PCa patient population.
| Supplementary Material | ▴Top |
Suppl 1. List of tested genes.
Suppl 2. Details of pathogenic/likely pathogenic/increased risk allele germline mutations.
Acknowledgments
None to declare.
Financial Disclosure
This project was funded by a competitive intramural grant from King Hussein Cancer Center.
Conflict of Interest
Edward D. Esplin and Sarah M. Nielsen are employees and shareholders of Invitae Corp. Brandie Heald are former employees and shareholders of Invitae Corp. Edward D. Esplin is an advisor and stockholder of Taproot Health and Exir Bio. All listed authors have no conflict of interest to declare.
Informed Consent
All enrolled patients signed consent forms.
Author Contributions
Conceptualization: HAR. Data curation: RAH, SS, RG, AA, HBH. Formal analysis: HAR, HBH, RAH, SS. Investigation: SMN, BS, HBH. Project administration: HAR, SMN, HBH. Supervision: HAR, HBH, RAH. Writing-original draft: RAH, HAR. Writing-review and editing: HAR, RAH, BS, SMN, BH, EDS, HBH, SS, RG, AA, TA, YA.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request. All variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
| References | ▴Top |
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229-263.
doi pubmed - Turco F, Tucci M, Delcuratolo MD, Di Stefano RF, Pisano C, Audisio A, Audisio M, et al. Treatment intensification for metastatic prostate cancer: new treatment landscapes in androgen deprivation-based therapy. Cancer Commun (Lond). 2022;42(8):683-688.
doi pubmed pmc - Ong S, O'Brien J, Medhurst E, Lawrentschuk N, Murphy D, Azad A. Current treatment options for newly diagnosed metastatic hormone-sensitive prostate cancer-a narrative review. Transl Androl Urol. 2021;10(10):3918-3930.
doi pubmed pmc - Ciccarese C, Iacovelli R, Sternberg CN, Gillessen S, Tortora G, Fizazi K. Triplet therapy with androgen deprivation, docetaxel, and androgen receptor signalling inhibitors in metastatic castration-sensitive prostate cancer: a meta-analysis. Eur J Cancer. 2022;173:276-284.
doi pubmed - Gebrael G, Thomas VM, Swami U, Agarwal N. The management of metastatic castrate-sensitive prostate cancer: from guidelines to real-world practice. Oncologist. 2023;28(9):746-749.
doi pubmed pmc - Goh CL, Schumacher FR, Easton D, Muir K, Henderson B, Kote-Jarai Z, Eeles RA. Genetic variants associated with predisposition to prostate cancer and potential clinical implications. J Intern Med. 2012;271(4):353-365.
doi pubmed - Benafif S, Kote-Jarai Z, Eeles RA, Consortium P. A review of prostate cancer genome-wide association studies (GWAS). Cancer Epidemiol Biomarkers Prev. 2018;27(8):845-857.
doi pubmed pmc - Marino F, Totaro A, Gandi C, Bientinesi R, Moretto S, Gavi F, Pierconti F, et al. Germline mutations in prostate cancer: a systematic review of the evidence for personalized medicine. Prostate Cancer Prostatic Dis. 2023;26(4):655-664.
doi pubmed - Castro E, Romero-Laorden N, Del Pozo A, Lozano R, Medina A, Puente J, Piulats JM, et al. PROREPAIR-B: a prospective cohort study of the impact of germline DNA repair mutations on the outcomes of patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2019;37(6):490-503.
doi pubmed - Na R, Zheng SL, Han M, Yu H, Jiang D, Shah S, Ewing CM, et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol. 2017;71(5):740-747.
doi pubmed pmc - de la Calle CM, Bhanji Y, Pavlovich CP, Isaacs WB. The role of genetic testing in prostate cancer screening, diagnosis, and treatment. Curr Opin Oncol. 2022;34(3):212-218.
doi pubmed - Szymaniak BM, Facchini LA, Giri VN, Antonarakis ES, Beer TM, Carlo MI, Danila DC, et al. Practical considerations and challenges for germline genetic testing in patients with prostate cancer: recommendations from the Germline Genetics Working Group of the PCCTC. JCO Oncol Pract. 2020;16(12):811-819.
doi pubmed pmc - National Comprehensive Cancer Network (NCCN) Prostate Cancer Guidelines Version 3.2022, 2022. Available at: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf. (accessed on October 15, 2023).
- Giri VN, Knudsen KE, Kelly WK, Cheng HH, Cooney KA, Cookson MS, Dahut W, et al. Implementation of germline testing for prostate cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol. 2020;38(24):2798-2811.
doi pubmed pmc - Sanda MG, Cadeddu JA, Kirkby E, Chen RC, Crispino T, Fontanarosa J, Freedland SJ, et al. Clinically localized prostate cancer: AUA/ASTRO/SUO Guideline. Part I: risk stratification, shared decision making, and care options. J Urol. 2018;199(3):683-690.
doi pubmed - Morote J, Aguilar A, Planas J, Trilla E. Definition of castrate resistant prostate cancer: new insights. Biomedicines. 2022;10(3):689.
doi pubmed pmc - Lincoln SE, Kobayashi Y, Anderson MJ, Yang S, Desmond AJ, Mills MA, Nilsen GB, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17(5):533-544.
doi pubmed - Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, Kobayashi Y, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19(10):1105-1117.
doi pubmed pmc - Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, Garofalo A, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453.
doi pubmed pmc - Zhu Y, Wei Y, Zeng H, Li Y, Ng CF, Zhou F, He C, et al. Inherited mutations in Chinese men with prostate cancer. J Natl Compr Canc Netw. 2021;20(1):54-62.
doi pubmed - Giri VN, Hartman R, Pritzlaff M, Horton C, Keith SW. Germline variant spectrum among African American men undergoing prostate cancer germline testing: need for equity in genetic testing. JCO Precis Oncol. 2022;6(1):e2200234.
doi pubmed pmc - Fan C, Zhang J, Ouyang T, Li J, Wang T, Fan Z, Fan T, et al. RAD50 germline mutations are associated with poor survival in BRCA1/2-negative breast cancer patients. Int J Cancer. 2018;143(8):1935-1942.
doi pubmed - Zuntini R, Bonora E, Pradella LM, Amato LB, Vidone M, De Fanti S, Catucci I, et al. Detecting variants in the NBN gene while testing for hereditary breast cancer: what to do next? Int J Mol Sci. 2021;22(11):5832.
doi pubmed pmc - Forkosh E, Bergel M, Hatchell KE, Nielsen SM, Heald B, Benson AA, Friedman E, et al. Ashkenazi Jewish and other white APC I1307K carriers are at higher risk for multiple cancers. Cancers (Basel). 2022;14(23):5875.
doi pubmed pmc - Shore N, Gazi M, Pieczonka C, Heron S, Modh R, Cahn D, Belkoff LH, et al. Efficacy of National Comprehensive Cancer Network Guidelines in identifying pathogenic germline variants among unselected patients with prostate cancer: The PROCLAIM trial. Eur Urol Oncol. 2023;6(5):477-483.
doi pubmed - Briggs LG, Steele GL, Qian ZJ, Subbana S, Alkhatib KY, Labban M, Langbein BJ, et al. Racial differences in germline genetic testing for prostate cancer: a systematic review. JCO Oncol Pract. 2023;19(5):e784-e793.
doi pubmed - Tatineni S, Tarockoff M, Abdallah N, Purrington KS, Assad H, Reagle R, Petrucelli N, et al. Racial and ethnic variation in multigene panel testing in a cohort of BRCA1/2-negative individuals who had genetic testing in a large urban comprehensive cancer center. Cancer Med. 2022;11(6):1465-1473.
doi pubmed pmc - Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177(1):26-31.
doi pubmed pmc - So MK, Ahn HK, Huh J, Kim KH. Germline pathogenic variants in unselected Korean men with prostate cancer. Investig Clin Urol. 2022;63(3):294-300.
doi pubmed pmc - Gould D, Walker R, Makari-Judson G, Seven M. Experiences of individuals with a variant of uncertain significance on genetic testing for hereditary cancer risks: a mixed method systematic review. J Community Genet. 2022;13(4):371-379.
doi pubmed pmc - Mersch J, Brown N, Pirzadeh-Miller S, Mundt E, Cox HC, Brown K, Aston M, et al. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA. 2018;320(12):1266-1274.
doi pubmed pmc - Esterling L, Wijayatunge R, Brown K, Morris B, Hughes E, Pruss D, Manley S, et al. Impact of a cancer gene variant reclassification program over a 20-year period. JCO Precis Oncol. 2020;4:PO.20.00020.
doi pubmed pmc - Nicolosi P, Heald B, Esplin ED. What is a variant of uncertain significance in genetic testing? Eur Urol Focus. 2022;8(3):654-656.
doi pubmed - de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, Chi KN, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091-2102.
doi pubmed - Fizazi K, Piulats JM, Reaume MN, Ostler P, McDermott R, Gingerich JR, Pintus E, et al. Rucaparib or Physician's choice in metastatic prostate cancer. N Engl J Med. 2023;388(8):719-732.
doi pubmed pmc - Hussain M, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, Chi KN, et al. Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med. 2020;383(24):2345-2357.
doi pubmed - FDA approves olaparib with abiraterone and prednisone (or prednisolone) for BRCA-mutated metastatic castration-resistant prostate cancer. FDA Approves Olaparib with Abiraterone and Prednisone. Available at: https://esmo.org/oncology-news/fda-approves-olaparib-with-abiraterone-and-prednisone-or-prednisolone-for-brca-mutated-metastatic-castration-resistant-prostate-cancer (accessed on October 15, 2023).
- Saad F, Clarke NW, Oya M, Shore N, Procopio G, Guedes JD, Arslan C, et al. Olaparib plus abiraterone versus placebo plus abiraterone in metastatic castration-resistant prostate cancer (PROpel): final prespecified overall survival results of a randomised, double-blind, phase 3 trial. Lancet Oncol. 2023;24(10):1094-1108.
doi pubmed - Chi KN, Rathkopf D, Smith MR, Efstathiou E, Attard G, Olmos D, Lee JY, et al. Niraparib and abiraterone acetate for metastatic castration-resistant prostate cancer. J Clin Oncol. 2023;41(18):3339-3351.
doi pubmed pmc - Smith MR, Scher HI, Sandhu S, Efstathiou E, Lara PN, Jr., Yu EY, George DJ, et al. Niraparib in patients with metastatic castration-resistant prostate cancer and DNA repair gene defects (GALAHAD): a multicentre, open-label, phase 2 trial. Lancet Oncol. 2022;23(3):362-373.
doi pubmed pmc - Chi KN, Sandhu S, Smith MR, Attard G, Saad M, Olmos D, Castro E, et al. Niraparib plus abiraterone acetate with prednisone in patients with metastatic castration-resistant prostate cancer and homologous recombination repair gene alterations: second interim analysis of the randomized phase III MAGNITUDE trial. Ann Oncol. 2023;34(9):772-782.
doi pubmed pmc - Antonarakis ES, Shaukat F, Isaacsson Velho P, Kaur H, Shenderov E, Pardoll DM, Lotan TL. Clinical features and therapeutic outcomes in men with advanced prostate cancer and DNA mismatch repair gene mutations. Eur Urol. 2019;75(3):378-382.
doi pubmed pmc - Isaacsson Velho P, Silberstein JL, Markowski MC, Luo J, Lotan TL, Isaacs WB, Antonarakis ES. Intraductal/ductal histology and lymphovascular invasion are associated with germline DNA-repair gene mutations in prostate cancer. Prostate. 2018;78(5):401-407.
doi pubmed pmc - Sedhom R, Antonarakis ES. Clinical implications of mismatch repair deficiency in prostate cancer. Future Oncol. 2019;15(20):2395-2411.
doi pubmed pmc - Schweizer MT, Cheng HH, Tretiakova MS, Vakar-Lopez F, Klemfuss N, Konnick EQ, Mostaghel EA, et al. Mismatch repair deficiency may be common in ductal adenocarcinoma of the prostate. Oncotarget. 2016;7(50):82504-82510.
doi pubmed pmc - Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, Rathkopf D, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 2019;5(4):471-478.
doi pubmed pmc - Graham LS, Montgomery B, Cheng HH, Yu EY, Nelson PS, Pritchard C, Erickson S, et al. Mismatch repair deficiency in metastatic prostate cancer: response to PD-1 blockade and standard therapies. PLoS One. 2020;15(5):e0233260.
doi pubmed pmc - Hansen AR, Massard C, Ott PA, Haas NB, Lopez JS, Ejadi S, Wallmark JM, et al. Pembrolizumab for advanced prostate adenocarcinoma: findings of the KEYNOTE-028 study. Ann Oncol. 2018;29(8):1807-1813.
doi pubmed - Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019;25(13):3753-3758.
doi pubmed - Paller CJ, Antonarakis ES, Beer TM, Borno HT, Carlo MI, George DJ, Graff JN, et al. Germline genetic testing in advanced prostate cancer; practices and barriers: survey results from the Germline Genetics Working Group of the Prostate Cancer Clinical Trials Consortium. Clin Genitourin Cancer. 2019;17(4):275-282.e271.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


