| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 5, October 2024, pages 792-800
Analysis of Pancreatic Cancer Genetic Risk Factors in a Multi-Ethnic Population Sample
Abdullah Al-Qahtania, Ali Al-Alib, c, Bency Johnd, Kusum Kapilad, e, Rabeah Al-Temaimid, f
aUndergraduate Medical Program, College of Medicine, Kuwait University, Jabriya, Kuwait
bDepartment of Gastroenterology and Hepatology, Mubarak Al Kabeer Hospital, Jabriya, Kuwait
cDepartment of Medicine, College of Medicine, Kuwait University, Jabriya, Kuwait
dDepartment of Pathology, College of Medicine, Kuwait University, Jabriya, Kuwait
eDepartment of Laboratory Medicine, Mubarak Al Kabeer Hospital, Jabriya, Kuwait
fCorresponding Author: Rabeah Al-Temaimi, Department of Pathology, College of Medicine, Kuwait University, Jabriya, Kuwait
Manuscript submitted June 17, 2024, accepted August 1, 2024, published online August 10, 2024
Short title: PDAC Genetic Risk
doi: https://doi.org/10.14740/wjon1911
| Abstract | ▴Top |
Background: Pancreatic cancer (PC) has one of the highest mortality to incidence ratio of all cancers. Early identification of at-risk individuals should permit early diagnosis. Genome-wide association studies showed the association of several genetic variants with PC risk in multi-ethnic populations. Our objective was to examine the association of these genetic variants with PC in a population sample from Kuwait.
Methods: DNA samples from 103 pancreatic ductal adenocarcinoma (PDAC) specimens and 132 healthy controls were used for genotyping ABO rs505922, BCAR1 rs7190458, LINC-PINT rs6971499, HNF1B rs4795218, VDR rs2228570 rs731236, and PRSS1 rs111033565 rs111033568 rs387906698 and rs267606982 using TaqMan genotyping assays, and VDR expression was performed by immunocytochemistry.
Results: ABO rs505922C and VDR rs2228570A were associated with PDAC risk (odds ratio (OR): 1.55, 95% confidence interval (CI): 1.07 - 2.24, P = 0.027; OR: 1.64, 95% CI: 1.09 - 2.48, P = 0.024; respectively). An unweighted polygenic risk score (ABO rs505922, BCAR1 rs7190458, LINC-PINT rs6971499, and HNF1B rs4795218) was significantly associated with PDAC risk (β: -0.11, 95% CI: -0.15 to -0.05, P < 0.001). VDR expression was downregulated or absent in most PDAC specimens regardless of VDR haplotype.
Conclusion: ABO rs505922C and VDR rs2228570A are PDAC genetic risk factors in our population. Ethnicity influences the association of reported genetic PDAC risk factors and should be adjusted for when performing PDAC genetic risk estimations. Investigation of these genetic risk factors in other ethnic populations is a necessity to evaluate their PDAC risk prediction potential.
Keywords: Pancreatic cancer; Pancreatic ductal adenocarcinoma; Genetic risk; Ethnicity; ABO; Vitamin D receptor
| Introduction | ▴Top |
Pancreatic cancer (PC) is one of the most aggressive types of cancer and ranks third in the mortality-to-incidence ratio of all cancers [1]. The 5-year survival rate for PC is still 9% despite advances in diagnostic and treatment protocols in the past decade [2]. As with other cancers, those diagnosed at the early stages of the disease and with localized or resectable small tumors have better odds of survival. In general, PC is rarely diagnosed at an early stage. The most commonly diagnosed type of PC is pancreatic adenocarcinoma or pancreatic ductal adenocarcinoma (PDAC), which accounts for 90% of all PC cases. Other rarer types include neuroendocrine PCs, adenosquamous carcinoma, squamous cell carcinoma, and colloid carcinoma.
The risk factors of PC can be grouped into modifiable and non-modifiable risk factors [2]. The modifiable risk factors are smoking, alcohol consumption, obesity, and chronic pancreatitis. The non-modifiable risk factors include age, incidence of diabetes, sex, ethnicity, blood group, infection, family history of PC, and inherited genetic defects. In non-hereditary PC, the majority of PC patients are 65 - 70 years of age; thus, it is considered a disease of the elderly. Studies showed that patients with blood groups A, AB, or B are at a higher risk of developing PDAC than those with blood group O [3]. A meta-analysis study showed that patients with type 1 diabetes have a twofold risk of developing PC than those without type 1 diabetes [4]. Another meta-analysis study demonstrated similar findings with type 2 diabetes patients [5]. In addition, patients with a family history of PC have a nine times higher risk of developing it [6]. Moreover, several hereditary cancer syndromes are associated with increases in PC risk, such as Peutz-Jeghers syndrome (100-fold), and familial atypical multiple mole melanoma syndromes (13- to 38-fold). However, these cancer syndromes are very rare and only account for 5-10% of all PCs [7]. Therefore, sporadic PC accounts for about 90% of all PCs, and identifying genetic PC risk factors is essential for early predicting and detecting the disease. In addition, mutations in PRSS1 predispose to familial pancreatitis, a known risk factor for PC. Moreover, several genome-wide association studies (GWASs) have attempted to identify these genetic risk factors in diverse populations [3, 8-11]. In total, 31 genetic risk factors have been reported to be associated with PC risk, of which 71% were specific to European-Caucasians. Since ethnicity contributes to differences in PC risk, assessing whether these risk factors can be applied to the global population is imperative to ascertain their predictive potential and perceived predictive application. A recent study on a multi-ethnic PC population attempted to assess whether the 31 GWAS-reported PC risk factors can be applied to an ethnically diverse population [12]. They reported 11 of the 31 genetic variants associated with PC risk in a multi-ethnic population sample at various degrees of significance, whereas ethnic-specific associations varied considerably. The variants with the highest degree of significance were ABO rs505922, BCAR1 rs7190458, LINC-PINT rs6971499, and HNF1B rs4795218, all of which maintained their association with PC risk in European-Caucasians, and the multi-ethnic PC population samples. The Kuwaiti population is multi-ethnic, and the Kuwaiti (Arab) genetic background aligns well with European ancestry [13]. Therefore, we prioritized these four variants to investigate their association with PC in a cohort sampled from Kuwait. In addition, vitamin D and its receptor polymorphisms have been shown to have a role in PC risk, pathogenesis, progression, response to treatment, and overall survival [14-17]. In this study, we aimed to assess 10 potential PC genetic risk variants, including four PRSS1 variants as they associate with the risk of cancer-predisposing pancreatitis, four GWAS variants, and two vitamin D receptor (VDR) variants in an admixed PC population sample from Kuwait. Moreover, we examined the expression patterns of VDR in PDAC under the influence of different VDR haplotypes.
| Materials and Methods | ▴Top |
Study cohorts
All study protocols were approved by Kuwait’s Ministry of Health ethical review committee, which adheres to the Declaration of Helsinki’s Ethical Principles for Medical Research Involving Human Subjects guidelines. Verbal consent was secured from participants undergoing biopsy procurement for assessment and was maintained for all archival cell block specimens included in this study. Healthy control participants included in this study provided informed written consent. A hundred and forty archival formalin-fixed paraffin-embedded (FFPE) fine-needle aspirate (FNA) cell blocks collected between 2016 and 2023 were assessed for suitability for downstream analyses. Patient inclusion criteria included having a patient file on record with demographics, final clinical diagnosis, and sample acquisition site. Exclusion criteria included patients with a PC diagnosis at an age younger than 45 years, as it may suggest an inherited genetic predisposition. Assessment criteria of hematoxylin and eosin (H&E)-stained cell block sections included having sufficient cell material, clearly identified cell structures, location of FNA sample (head, tail, or body of the pancreas), and having a confirmed final PC diagnosis. Exclusion criteria included insufficient cell material and having undefined cellular structures. One hundred and three PC specimens satisfied the inclusion criteria and were used in this study. A hundred and thirty-two healthy control DNA samples were retrieved from the laboratory biobank. Healthy control inclusion criteria were not having any chronic disease diagnosis and having complete demographic data. Exclusion criteria were a personal history of cancer, a diagnosis of diabetes in the family, and incomplete demographic data. Healthy controls older than 50 years of age were prioritized.
DNA extraction
Eight 10 µm thick sections were cut from each FNA cell block and mounted on slides. Normal cell infiltrates were ascertained for each specimen using an H&E-stained guide slide. Normal cells were macro dissected using a sterile 21-gauge needle and collected in a sterile 1.5 mL microcentrifuge containing xylene. Macro dissected specimens were deparaffinized in xylene at room temperature for 12 min, followed by centrifugation at 14,000 rpm for 5 min. The resultant pellet was washed twice with absolute ethanol and centrifuged at 14,000 rpm for 5 min. The final pellet was dried by incubation at 37 °C for 15 min. QIAmp DNA mini extraction kit was used for DNA extraction according to the manufacturer’s recommended protocol (Qiagen, MD, USA). In brief, the pellet was resuspended in 180 µL of ATL lysis solution and 20 µL of proteinase K. Samples were vortexed for 15 s and incubated overnight in an incubator shaker set at 200 rpm and 56 °C. After incubation, 200 µL of lysis buffer AL was added and incubated at 70 °C for 10 min. Absolute ethanol was added, and the sample was vortexed to mix. The final suspension was loaded into a spin filter column and centrifuged at 8,000 rpm for 1 min. The column was washed twice with 500 µL of wash buffers of different stringencies, followed by centrifugations at 8,000 rpm for 1 min and 14,000 rpm for 3 min, respectively. After the final wash, the column was spun at maximum speed to dry, and 50 µL of Tris-buffered saline (TBS) pH 8.5 was added to the column. DNA elution was performed by centrifugation at 8,000 rpm for 1 min. Extracts were assessed for quantity using a spectrophotometer.
Variant genotyping
Taqman genotyping assays (Thermo Fisher Scientific, Waltham, MA, USA) were used for all variants according to the manufacturer’s protocols for allelic discrimination experiments. The variants analyzed included ABO rs505922, BCAR1 rs7190458, LINC-PINT rs6971499, HNF1B rs4795218, VDR rs2228570 rs731236, and PRSS1 rs111033565 rs111033568 rs387906698 and rs267606982. In brief, 10 ng of DNA was added to a 20 µL reaction mixture containing 1X TaqMan Genotyping Master Mix and 1X probes/primers assay. Reactions were loaded in a 96-well plate and run on a QuantStudio 3 system (Life Technologies, Carlsbad, CA, USA) according to the preset run method for a genotyping experiment. Allelic discrimination analysis was performed on the instrument’s data analysis software.
VDR immunocytochemistry
PC FNA cell blocks were sectioned into 4 - 5 µm thin sections and stained with H&E to confirm cellularity. Sections of each cell block were deparaffinized in three changes of fresh xylene for 3 min each, followed by a descending alcohol series (100%, 90%, and 70% ethanol) hydration protocol. Sections were submerged into three changes of each ethanol concentration and incubated for 3 min. Sections were rinsed in deionized water, and antigen retrieval was performed using 0.01 M citrate buffer (pH 6.0) and microwave incubation for 20 min. Slides were cooled for 20 min, rinsed with deionized water, and incubated in TBS (pH 7.6) for 5 min at room temperature. Slides were blocked in serum-free protein block (Agilent, CA, USA) for 30 min at room temperature, followed by incubation with VDR rabbit polyclonal antibody (NBP2-98841H, Novus Biologicals, CO, USA) at 1:100 dilution in 1% goat serum-TBS overnight at 4 °C. Slides were rinsed the next day in three washes of TBS and incubated with EnVision+ peroxidase-conjugated anti-mouse antibody (Agilent, CA, USA) for 30 min at room temperature. Slides were washed in TBS for 5 min, blocked with 3% hydrogen peroxide for 10 min, rinsed with TBS, and incubated with diaminobenzidine (Agilent Technologies, Glostrup, Denmark) for 2 - 5 min. Slides were counterstained with hematoxylin for 30 s, washed in tap water, and dehydrated in ascending alcohol concentrations. The slides were cleared in three changes of xylene for 3 min each and mounted in dibutylphthalate polystyrene xylene.
Statistical analysis
All genetic variants were assessed for deviation from Hardy-Weinberg equilibrium. The Fisher’s exact test was used for allelic distribution and inheritance pattern modeling, and the Chi-square test was used for genotype distribution analysis. Simple allele count unweighted polygenic risk score (uwPRS) and effect size weighted polygenic risk score (wPRS) calculation were conducted [18]. For wPRS estimation, we used log odds ratio (OR) values from Bogumil et al [12]. Logistic regression was performed on uwPRS and wPRS to determine their performance metrics. A P-value less than 0.05 was considered significant for all analyses. All analyses were performed using the Statistical Package for Social Sciences (SPSS) v. 28 (SPSS Inc., IL, USA).
| Results | ▴Top |
PC genetic risk factors
Healthy controls included in this study had an average age of 56.86 ± 16.87 years, and females comprised 63.6% of the cohort. The PC patients’ cohort had an average age of 63.44 ± 11 years, and females comprised 33% of the cohort. All PC samples were PDACs with two foamy cell PDAC variants. The demographic and clinical features of PC samples and healthy controls used in this study are shown in Table 1. The PC cohort displayed the following features: males had a higher incidence of PC than females, the age range with the highest incidence of PC was 50 - 59 years (33.75% of cases), and most of the samples were late-stage PDAC (74.8%). The non-Kuwaiti PC subgroup comprised of the following nationalities: 50% Middle Eastern (Syrian, Jordanian, Lebanese, Iraqi, Saudi, Yemeni), 16.7% African (Egyptian, Eritrean), 14.8% South-Asian (Indian, Pakistani, Bangladeshi), 9.3% Southeast-Asian (Malaysian, Filipinos), 7.4% Iranian, and 1.8% British. The healthy controls non-Kuwaiti subgroup comprised 43.2% Middle Eastern, 18.2% South Asian, and 18.2% African nationalities.
 Click to view | Table 1. Demographic and Clinical Characteristics of Pancreatic Cancer and Healthy Control Cohorts |
The 10 variants were genotyped in all PC samples and healthy controls. All PRSS1 variants were wild-type and were excluded from the analysis. The remaining six variants’ allelic and genotype frequencies are shown in Table 2. The healthy control cohort genotype distributions for the six variants met with the Hardy-Weinberg equilibrium assumption.
 Click to view | Table 2. Allele and Genotype Frequencies of Six Variants Assessed in Pancreatic Cancer and Healthy Control Samples |
Only two variants showed allelic association with PDAC risk, ABO rs505922C (OR: 1.55, 95% confidence interval (CI): 1.07 - 2.24, P = 0.027) and VDR rs2228570A (OR: 1.64, 95% CI: 1.09 - 2.48, P = 0.024). Inheritance model analysis showed both variants; ABO rs505922C (OR: 2.56, 95% CI: 1.24 - 5.5, P = 0.022) and VDR rs2228570A (OR: 2.64, 95% CI: 1.08 - 6.25, P = 0.044) confer their PDAC risk in an autosomal recessive mode of inheritance.
PRS analysis excluded VDR rs2228570A log OR to avoid ethnic bias in variant effect size compared to other included variants’ effect sizes retrieved from Bogumil et al [11]. The mean uwPRS in the healthy control cohort (mean 1.4) was significantly lower than the PC cohort (mean: 1.65, β: -0.11, 95% CI: -0.15 to -0.05, P < 0.001). The uwPRS distribution remained significant after adjusting for age and sex (P < 0.001). Similarly, the mean wPRS significantly differed between the healthy controls (mean: 0.31) and PC cohort (mean: 0.42) (β: -0.29, 95% CI: -0.47 to -0.1, P = 0.003), which remained significant after adjusting for age and sex (P = 0.007). Using logistic regression, we assessed whether uwPRS and wPRS can accurately classify PC patients with and without age and sex factors (Table 3). Genetic risk factors alone failed to explain the variation in PC risk, though they have potential specificity. Moreover, the inclusion of sex and age as established risk factors for PC refined the test performance metrics of the PRS models but failed to reach acceptable performance metrics.
 Click to view | Table 3. Unweighted and Weighted PRS Models’ Test Performance Metrics With and Without Age and Sex Inclusion |
VDR expression in PC
The two VDR variants analyzed here impact VDR expression and stability. The effect of VDR variant genotypes and haplotypes on VDR expression in PDAC was ascertained in specimens of adequate cellularity for investigation. Most PDAC specimens showed weak positive nuclear staining (54%) or negative VDR expression (40%). Only a few had weak cytoplasmic staining (4%) or cytoplasmic and nuclear staining (2%). VDR haplotypes did not correlate with VDR expression patterns (Fig. 1); however, generally, VDR expression was either suppressed or confined to the nucleus at low levels without evidence of translocation to the plasma membrane or the cytoplasm.
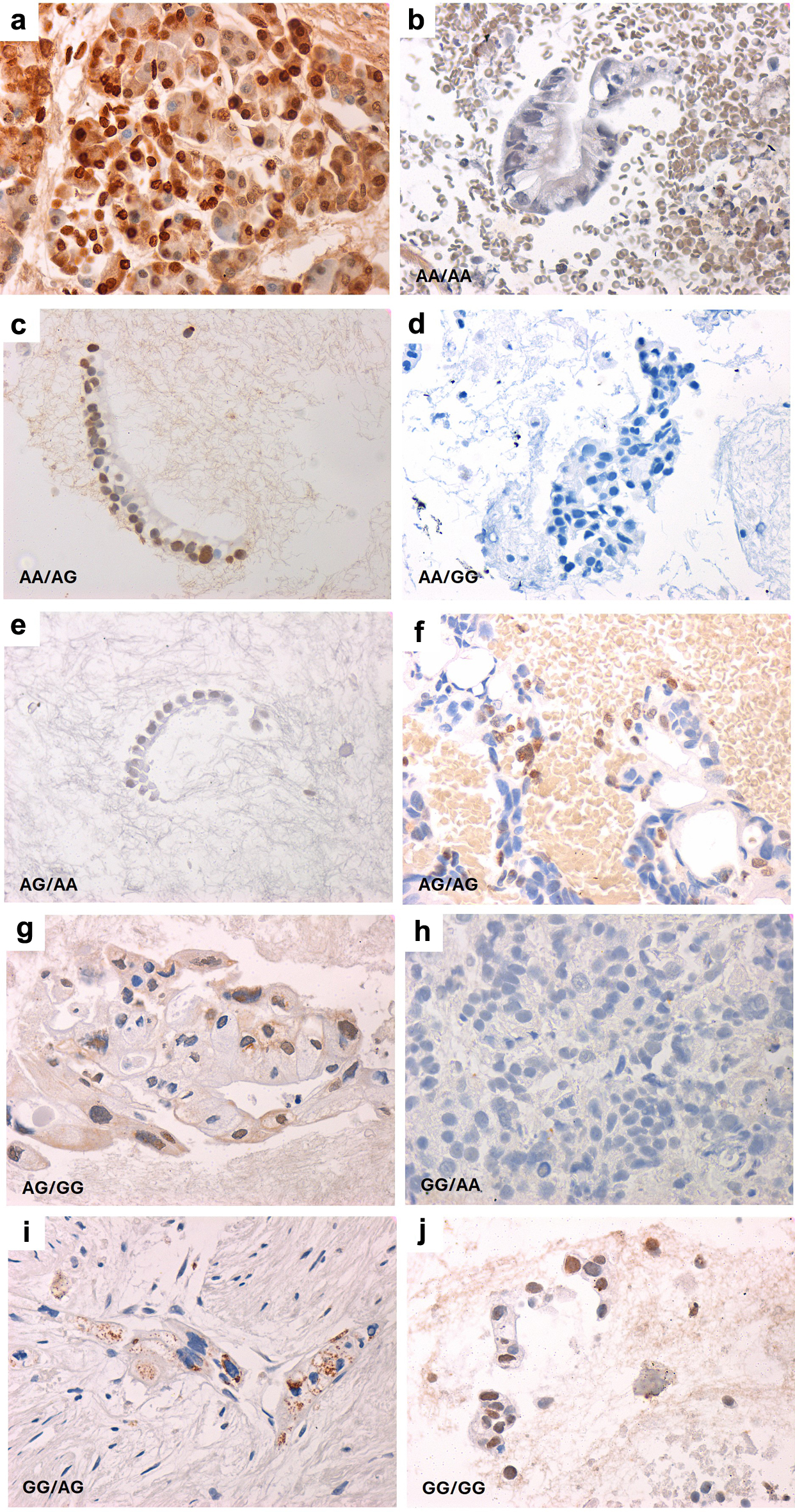 Click for large image | Figure 1. Representative images of VDR expression in pancreatic cancer according to VDR variants’ haplotype. VDR haplotypes are shown as rs2228570/rs731236. (a) Normal VDR expression in a healthy pancreatic specimen. (b-j) Representative images of pancreatic cancer cell block sections according to VDR haplotypes. All images were captured at × 400 magnification. |
| Discussion | ▴Top |
The genetic risk contribution to sporadic PC incidence has eluded researchers for the past two decades. With the development of genome-wide association technologies, many genetic risk factors were reported, mostly from European and East Asian populations [9, 19-22]. Only a few studies addressed whether these genetic risk factors can be extended to other ethnic or multi-ethnic populations [12, 23]. Here, we selected PC genetic risk variants validated in multi-ethnic and European populations. We assessed their association with PC risk in a multi-ethnic cohort comprising nationalities that are predominantly from West-Asian countries. The four GWAS discovered variants assessed here were all intronic variants, except for BCAR1 rs7190458, a transition-synonymous variant. Only the ABO rs505922C allele sustained its association with PC risk in our multi-ethnic population sample. This variant is associated with having a blood group other than the O group. It has also been shown to be associated with the risk of developing many diseases, including cancer and venous thrombosis [24, 25]. The lack of association of BCAR1 rs7190458, LINC-PINT rs6971499, and HNF1B rs4795218 with PC risk in our multi-ethnic population sample may indicate that the populations from which these were reported did not include West-Asian ethnicity, which is the ethnicity most represented in our cohort. When we compared variant frequencies of our PC cohort to the 1000 genome frequencies (Supplementary Material 1, www.wjon.org), which is most represented with European ethnicity, BCAR1 rs7190458, and HNF1B rs4795218 were associated with PC risk in our cohort (P < 0.002). These findings suggest that ethnicity does influence the association of these variants with PC risk [26]. Furthermore, using these variants in a PRS computed by allele count (uwPRS) showed better performance metrics in our cohort than using effect sizes generated from another multi-ethnic population sample. While the OR increased, the degree of significance decreased along with test prediction and sensitivity metrics. In addition, including age and sex enhanced the PRS performance metrics except for test specificity. These findings suggest the need to validate PRSs across multi-ethnic populations and to include non-genetic risk factors to better determine their applicability in the early prediction of PC risk [27].
We considered VDR rs2228570 and rs731236 variants for their association with PC risk because they were shown previously to associate variably with cancer [28]. VDR rs2228570 allele A is this variant’s minor and ancestral allele and is in 5’ initiator codon sequence of the gene. The impact of this change results in reduced transcription of the VDR gene and a longer VDR protein compared to the wild-type G allele. VDR rs731236 is a synonymous variant for which the major allele A is associated with higher VDR expression. In our study, VDR rs2228570A was moderately associated with PC risk. Previously reported multi-ethnic meta-analyses showed a similar association for this variant with PC risk as well as risk for other cancers [29, 30]. However, rs2228570A has different frequencies in different ethnic populations, with Asian ethnicity having the highest frequency (0.43) and African ethnicity having the lowest (0.27). Therefore, our result may be limited to our ethnic population. Nevertheless, we examined the influence of rs2228570 on VDR expression patterns in PDAC. We found no significant impact for this variant alone or combined with rs731236. Moreover, we found VDR to be downregulated or absent in most samples and mainly confined to the nucleus. This suggests that signaling pathways of VDR are disrupted in PC, and the proposed benefits of vitamin D supplementation in PDAC patients may have merit [31, 32].
The limitations of our study include a small sample size, a low representation of some ethnicities, and a limited number of genetic factors assessed. However, given that PC incidence in Kuwait is 4.2 per 100,000 individuals in a population of 4.269 million [33], and the presence of eight government hospitals, a theoretical estimate of 157 PC cases per hospital is expected within the time frame of our study (2016 - 2023). We had 140 specimens on record of which 103 met the inclusion criteria of our study. Moreover, our inclusion criteria did not discriminate by ethnicity, which has contributed to the low presentation of some ethnicities. Lastly, the rationale for selecting genetic risk factors to be assessed in this study was rigorously scrutinized and the resultant risk factors assessed here are backed with reported evidence with limited bias. An advantage of our study is that it is the first study to report the association of genetic risk factors with PC risk in Kuwait specifically and in the region generally. In addition, we have shown that ABO rs505922 association with PC risk is not affected by ethnicity.
In conclusion, we have shown that ABO rs505922C and VDR rs2228570A are associated with PC risk in our population. Our findings highlighted the influence of ethnicity on the validity of reported PC genetic risk factors. It is necessary to validate these variants and their associated PRSs in diverse multi-ethnic populations to determine their potential use in the early prediction of PC risk.
| Supplementary Material | ▴Top |
Suppl 1. List of reported pancreatic cancer genetic risk factors from genome-wide association studies and their allele frequencies in different populations sourced from NCBI’s dbSNP 1000 genome data.
Acknowledgments
None to declare.
Financial Disclosure
Kuwait University funded the research leading to these results under grant agreement No. MG01/21.
Conflict of Interest
The authors have no conflict of interest to declare that are relevant to the content of this article.
Informed Consent
Participating patients provided informed verbal consent, whereas healthy volunteers provided informed written consent.
Author Contributions
The study conception and design were created by Rabeah Al-Temaimi. Kusum Kapila and Ali Al-Ali provided patient specimens and clinical data. Abdullah Al-Qahtani extracted DNA and performed genotyping experiments. Bency John performed cell block sectioning and immunohistochemistry. Kusum Kapila performed imaging. Rabeah Al-Temaimi performed data collection and statistical analysis. The manuscript was written by Rabeah Al-Temaimi. All authors read and approved the final manuscript.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
Abbreviations
BCAR1: breast cancer anti-estrogen resistance protein 1; DNA: deoxyribonucleic acid; FFPE: formalin-fixed paraffin-embedded; FNA: fine needle aspirate; GWAS: Genome-Wide Association Study; H&E: hematoxylin and eosin; HNF1B: hepatocyte nuclear factor-1 beta; LINC-PINT: long intergenic non-protein coding RNA, P53 induced transcript; PC: pancreatic cancer; PDAC: pancreatic ductal adenocarcinoma; PRS: polygenic risk score; PRSS1: serine protease 1; TBS: Tris-buffered saline; uwPRS: unweighted polygenic risk score; VDR: vitamin D receptor; wPRS: weighted polygenic risk score
| References | ▴Top |
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249.
doi pubmed - Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10(1):10-27.
doi pubmed pmc - Campa D, Gentiluomo M, Obazee O, Ballerini A, Vodickova L, Hegyi P, Soucek P, et al. Genome-wide association study identifies an early onset pancreatic cancer risk locus. Int J Cancer. 2020;147(8):2065-2074.
doi pubmed - Stevens RJ, Roddam AW, Beral V. Pancreatic cancer in type 1 and young-onset diabetes: systematic review and meta-analysis. Br J Cancer. 2007;96(3):507-509.
doi pubmed pmc - Huxley R, Ansary-Moghaddam A, Berrington de Gonzalez A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer. 2005;92(11):2076-2083.
doi pubmed pmc - McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24(43):4846-4861.
doi pubmed pmc - Klatte DCF, Wallace MB, Lohr M, Bruno MJ, van Leerdam ME. Hereditary pancreatic cancer. Best Pract Res Clin Gastroenterol. 2022;58-59:101783.
doi pubmed - Tang H, Wei P, Chang P, Li Y, Yan D, Liu C, Hassan M, et al. Genetic polymorphisms associated with pancreatic cancer survival: a genome-wide association study. Int J Cancer. 2017;141(4):678-686.
doi pubmed pmc - Klein AP, Wolpin BM, Risch HA, Stolzenberg-Solomon RZ, Mocci E, Zhang M, Canzian F, et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun. 2018;9(1):556.
doi pubmed pmc - Grant RC, Denroche RE, Borgida A, Virtanen C, Cook N, Smith AL, Connor AA, et al. Exome-wide association study of pancreatic cancer risk. Gastroenterology. 2018;154(3):719-722.e713.
doi pubmed pmc - Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, Bueno-de-Mesquita HB, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986-990.
doi pubmed pmc - Bogumil D, Conti DV, Sheng X, Xia L, Shu XO, Pandol SJ, Blot WJ, et al. Replication and genetic risk score analysis for pancreatic cancer in a diverse multiethnic population. Cancer Epidemiol Biomarkers Prev. 2020;29(12):2686-2692.
doi pubmed pmc - Alsmadi O, Thareja G, Alkayal F, Rajagopalan R, John SE, Hebbar P, Behbehani K, et al. Genetic substructure of Kuwaiti population reveals migration history. PLoS One. 2013;8(9):e74913.
doi pubmed pmc - Wei D, Wang L, Zuo X, Bresalier RS. Vitamin D: promises on the horizon and challenges ahead for fighting pancreatic cancer. Cancers (Basel). 2021;13(11):2716.
doi pubmed pmc - Li H, Ruan Y, Liu C, Fan X, Yao Y, Dai Y, Song Y, et al. VDR promotes pancreatic cancer progression in vivo by activating CCL20-mediated M2 polarization of tumor associated macrophage. Cell Commun Signal. 2024;22(1):224.
doi pubmed pmc - Li Z, Jia Z, Gao Y, Xie D, Wei D, Cui J, Mishra L, et al. Activation of vitamin D receptor signaling downregulates the expression of nuclear FOXM1 protein and suppresses pancreatic cancer cell stemness. Clin Cancer Res. 2015;21(4):844-853.
doi pubmed pmc - Innocenti F, Owzar K, Jiang C, Etheridge AS, Gordan R, Sibley AB, Mulkey F, et al. The vitamin D receptor gene as a determinant of survival in pancreatic cancer patients: Genomic analysis and experimental validation. PLoS One. 2018;13(8):e0202272.
doi pubmed pmc - Che R, Motsinger-Reif AA. Evaluation of genetic risk score models in the presence of interaction and linkage disequilibrium. Front Genet. 2013;4:138.
doi pubmed pmc - Wu C, Miao X, Huang L, Che X, Jiang G, Yu D, Yang X, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2011;44(1):62-66.
doi pubmed - Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan AA, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46(9):994-1000.
doi pubmed pmc - Ueno M, Ohkawa S, Morimoto M, Ishii H, Matsuyama M, Kuruma S, Egawa N, et al. Genome-wide association study-identified SNPs (rs3790844, rs3790843) in the NR5A2 gene and risk of pancreatic cancer in Japanese. Sci Rep. 2015;5:17018.
doi pubmed pmc - Lin Y, Nakatochi M, Hosono Y, Ito H, Kamatani Y, Inoko A, Sakamoto H, et al. Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nat Commun. 2020;11(1):3175.
doi pubmed pmc - Campa D, Rizzato C, Bauer AS, Werner J, Capurso G, Costello E, Talar-Wojnarowska R, et al. Lack of replication of seven pancreatic cancer susceptibility loci identified in two Asian populations. Cancer Epidemiol Biomarkers Prev. 2013;22(2):320-323.
doi pubmed - Duan YF, Zhu F, Li XD, An Y, Zhang H, Zhou Y, Zhang X, et al. Association between ABO gene polymorphism (rs505922) and cancer risk: a meta-analysis. Tumour Biol. 2015;36(7):5081-5087.
doi pubmed - Germain M, Saut N, Greliche N, Dina C, Lambert JC, Perret C, Cohen W, et al. Genetics of venous thrombosis: insights from a new genome wide association study. PLoS One. 2011;6(9):e25581.
doi pubmed pmc - Liew SZH, Ng KW, Ishak NDB, Lee SY, Zhang Z, Chiang J, Ngeow JYY. Geographical, ethnic, and genetic differences in pancreatic cancer predisposition. Chin Clin Oncol. 2023;12(3):27.
doi pubmed - Junior HLR, Novaes LAC, Datorre JG, Moreno DA, Reis RM. Role of polygenic risk score in cancer precision medicine of non-european populations: a systematic review. Curr Oncol. 2022;29(8):5517-5530.
doi pubmed pmc - Gnagnarella P, Raimondi S, Aristarco V, Johansson HA, Bellerba F, Corso F, Gandini S. Vitamin D receptor polymorphisms and cancer. Adv Exp Med Biol. 2020;1268:53-114.
doi pubmed - Ye ZM, Li LJ, Luo MB, Qing HY, Zheng JH, Zhang C, Lu YX, et al. A systematic review and network meta-analysis of single nucleotide polymorphisms associated with pancreatic cancer risk. Aging (Albany NY). 2020;12(24):25256-25274.
doi pubmed pmc - Deuster E, Jeschke U, Ye Y, Mahner S, Czogalla B. Vitamin D and VDR in gynecological cancers - a systematic review. Int J Mol Sci. 2017;18(11):2328.
doi pubmed pmc - Moz S, Contran N, Facco M, Trimarco V, Plebani M, Basso D. Vitamin D prevents pancreatic cancer-induced apoptosis signaling of inflammatory cells. Biomolecules. 2020;10(7):1055.
doi pubmed pmc - Wei D, Wang L, Liu Y, Hafley MA, Tan L, Lorenzi PL, Yang P, et al. Activation of vitamin D/VDR signaling reverses gemcitabine resistance of pancreatic cancer cells through inhibition of MUC1 expression. Dig Dis Sci. 2023;68(7):3043-3058.
doi pubmed - Elwali NE, AlShareef SM, Khamis AH, Elhassan MMA. Pancreatic cancer in Saudi Arabia (2005-2020): increasing trend. BMC Cancer. 2024;24(1):653.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


